Summary information and primary citation
- PDB-id
-
8xbw;
SNAP-derived features in text and
JSON formats
- Class
- DNA binding protein-DNA
- Method
- cryo-EM (2.89 Å)
- Summary
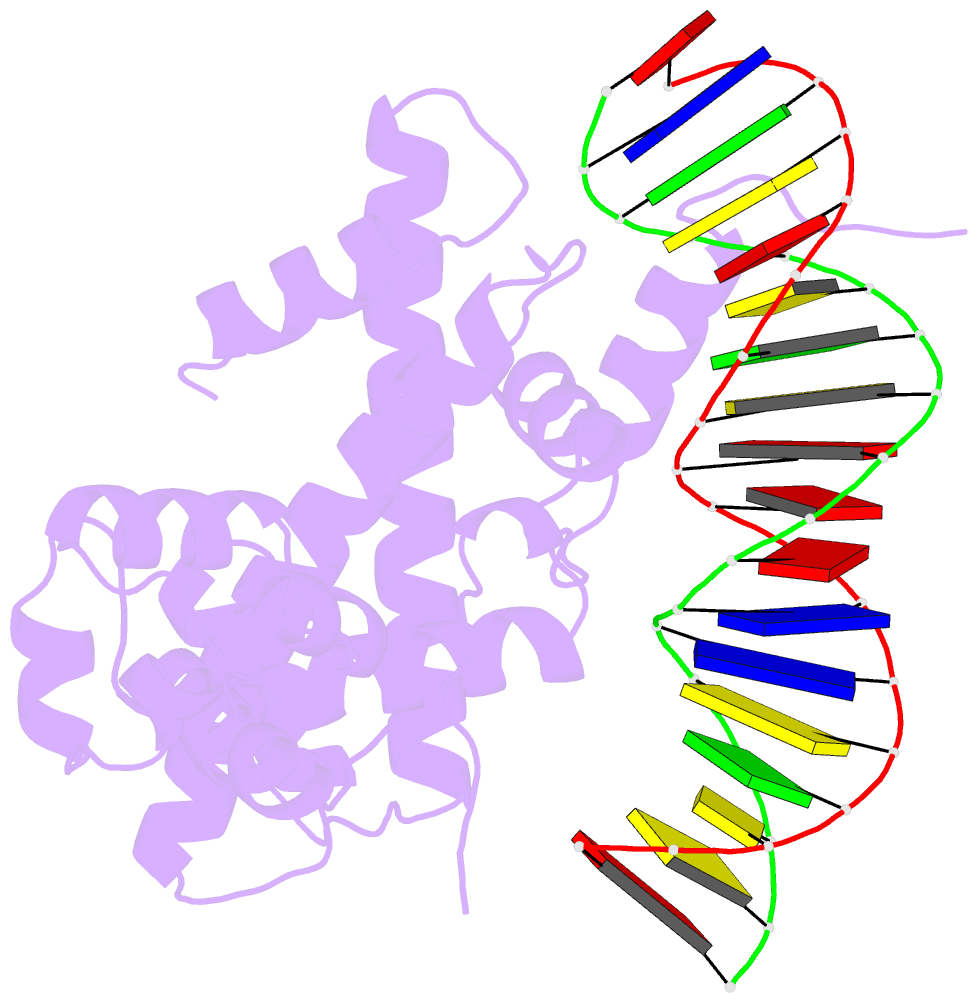
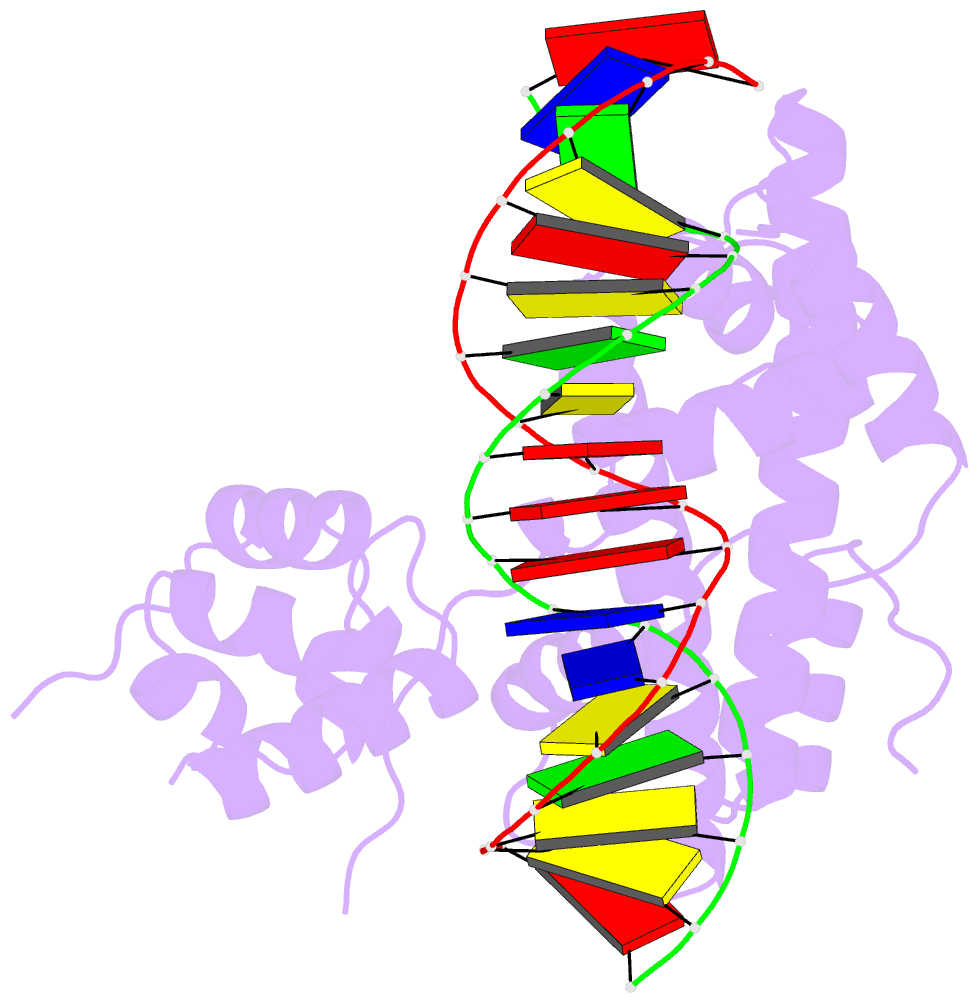
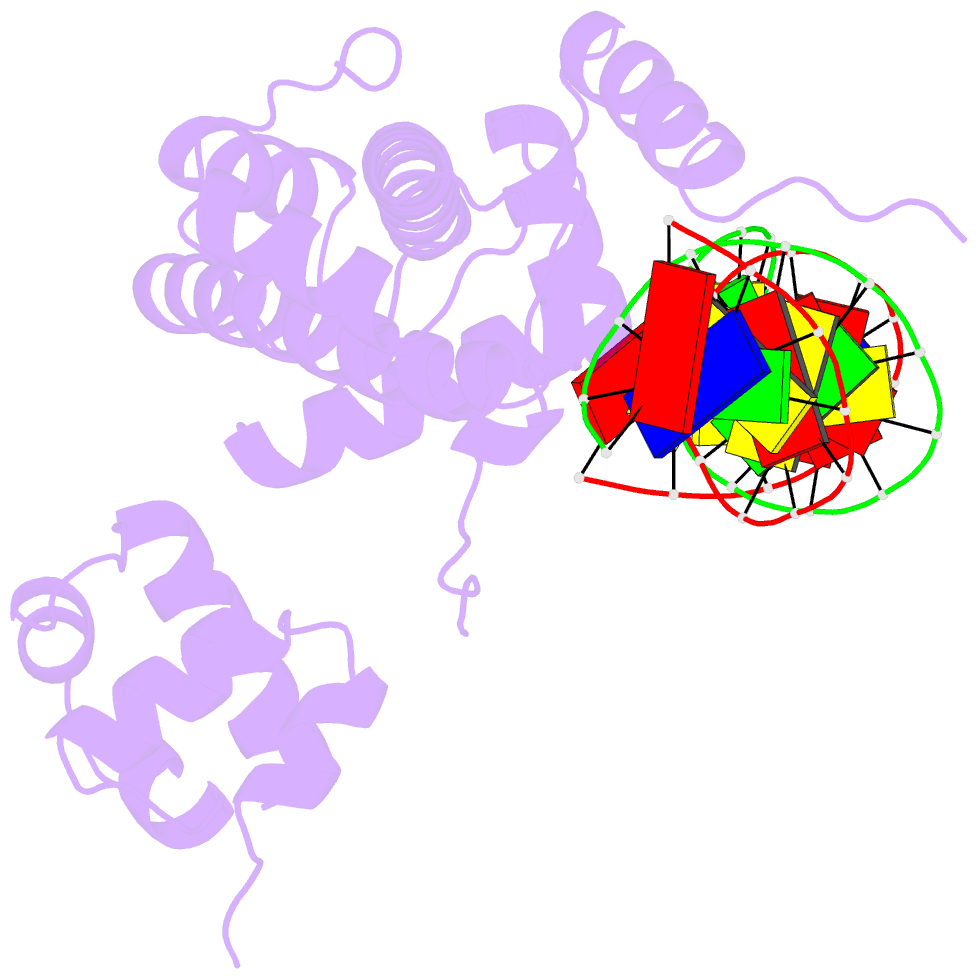
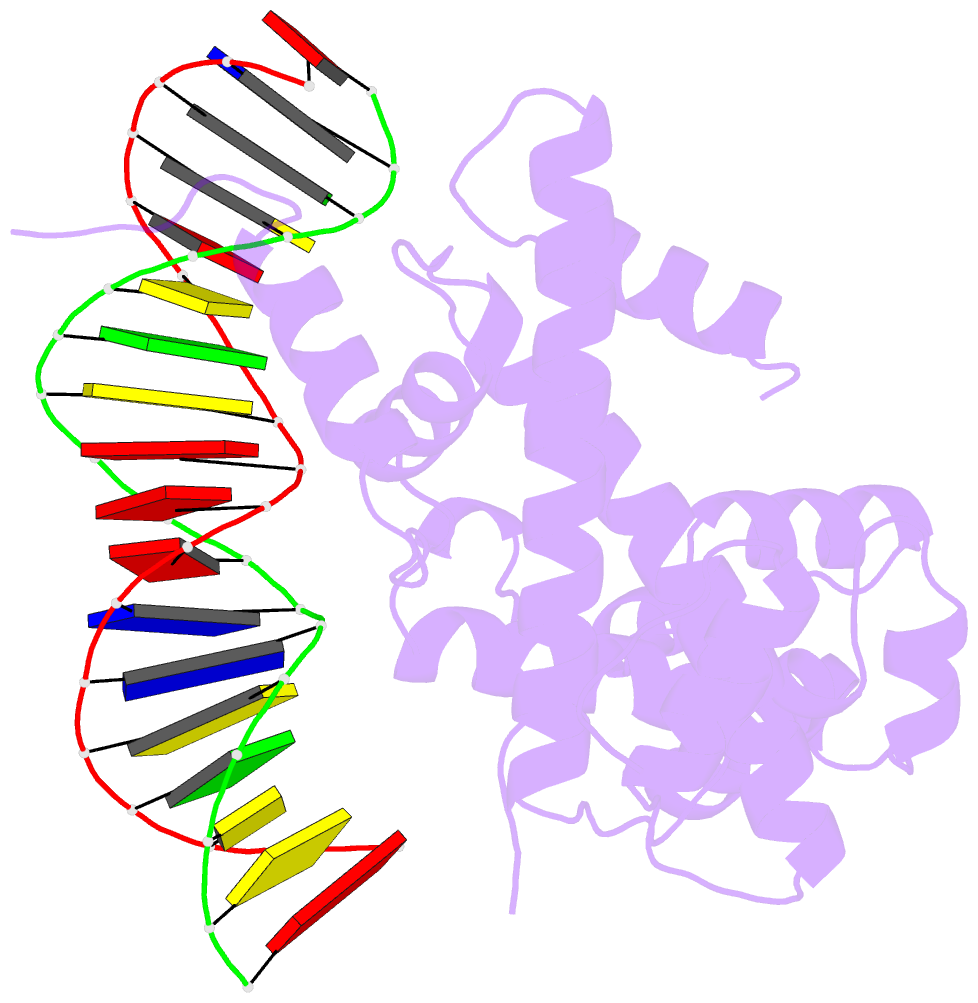
- The cryo-EM structure of the rad51 n-terminal lobe
domain bound to the histone h4 tail of the nucleosome
- Reference
-
Shioi T, Hatazawa S, Oya E, Hosoya N, Kobayashi W,
Ogasawara M, Kobayashi T, Takizawa Y, Kurumizaka H
(2024): "Cryo-EM
structures of RAD51 assembled on nucleosomes containing a
DSB site." Nature, 628,
212-220. doi: 10.1038/s41586-024-07196-4.
- Abstract
- RAD51 is the central eukaryotic recombinase required
for meiotic recombination and mitotic repair of
double-strand DNA breaks (DSBs)<sub>1,2</sub>.
However, the mechanism by which RAD51 functions at DSB
sites in chromatin has remained elusive. Here we report the
cryo-electron microscopy structures of human
RAD51-nucleosome complexes, in which RAD51 forms ring and
filament conformations. In the ring forms, the N-terminal
lobe domains (NLDs) of RAD51 protomers are aligned on the
outside of the RAD51 ring, and directly bind to the
nucleosomal DNA. The nucleosomal linker DNA that contains
the DSB site is recognized by the L1 and L2 loops-active
centres that face the central hole of the RAD51 ring. In
the filament form, the nucleosomal DNA is peeled by the
RAD51 filament extension, and the NLDs of RAD51 protomers
proximal to the nucleosome bind to the remaining
nucleosomal DNA and histones. Mutations that affect
nucleosome-binding residues of the RAD51 NLD decrease
nucleosome binding, but barely affect DNA binding in vitro.
Consistently, yeast Rad51 mutants with the corresponding
mutations are substantially defective in DNA repair in
vivo. These results reveal an unexpected function of the
RAD51 NLD, and explain the mechanism by which RAD51
associates with nucleosomes, recognizes DSBs and forms the
active filament in chromatin.