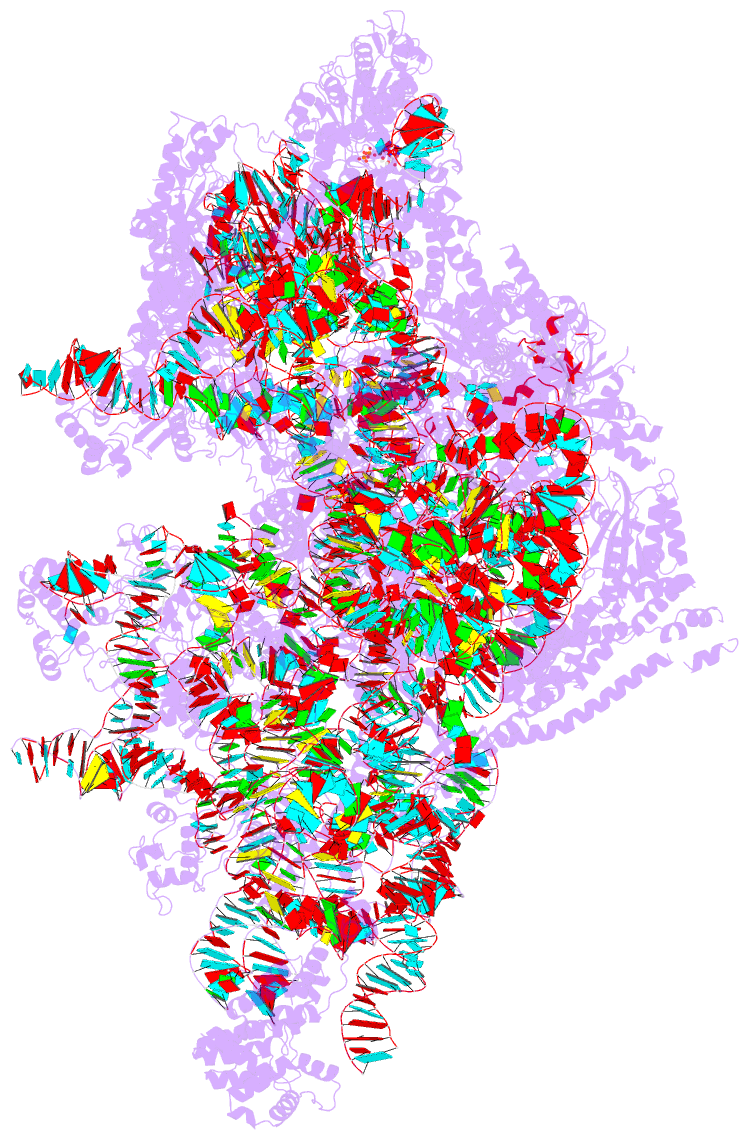
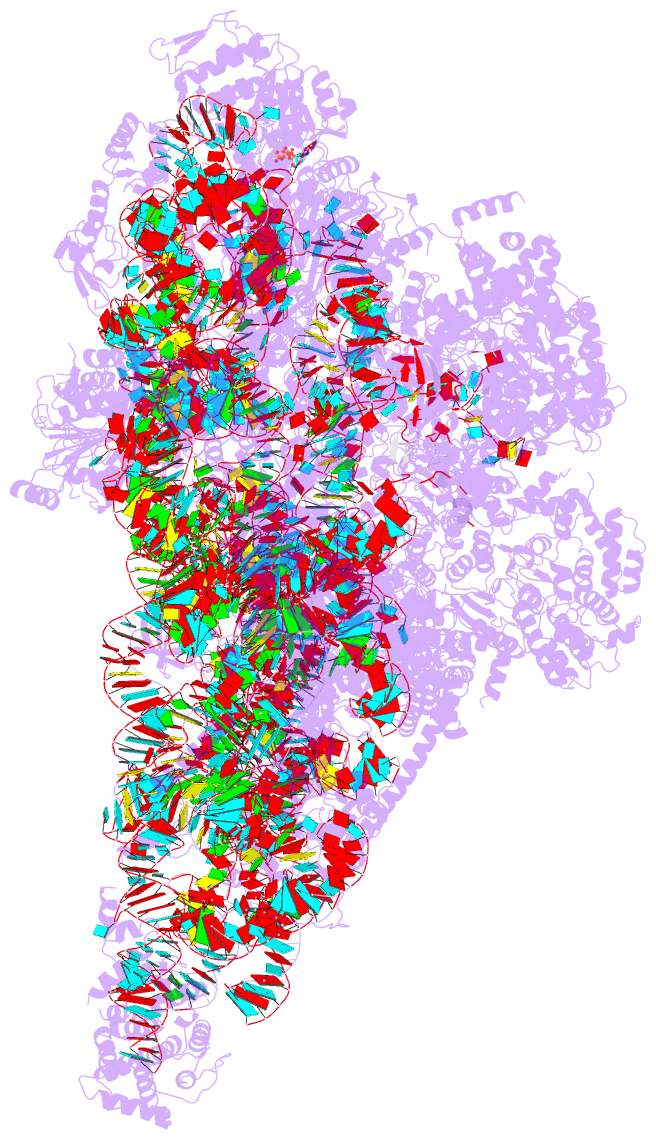
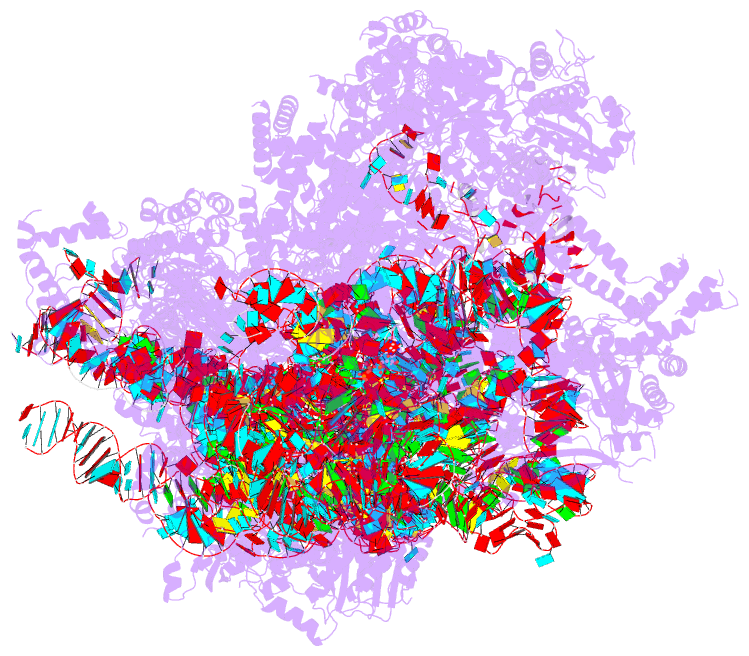
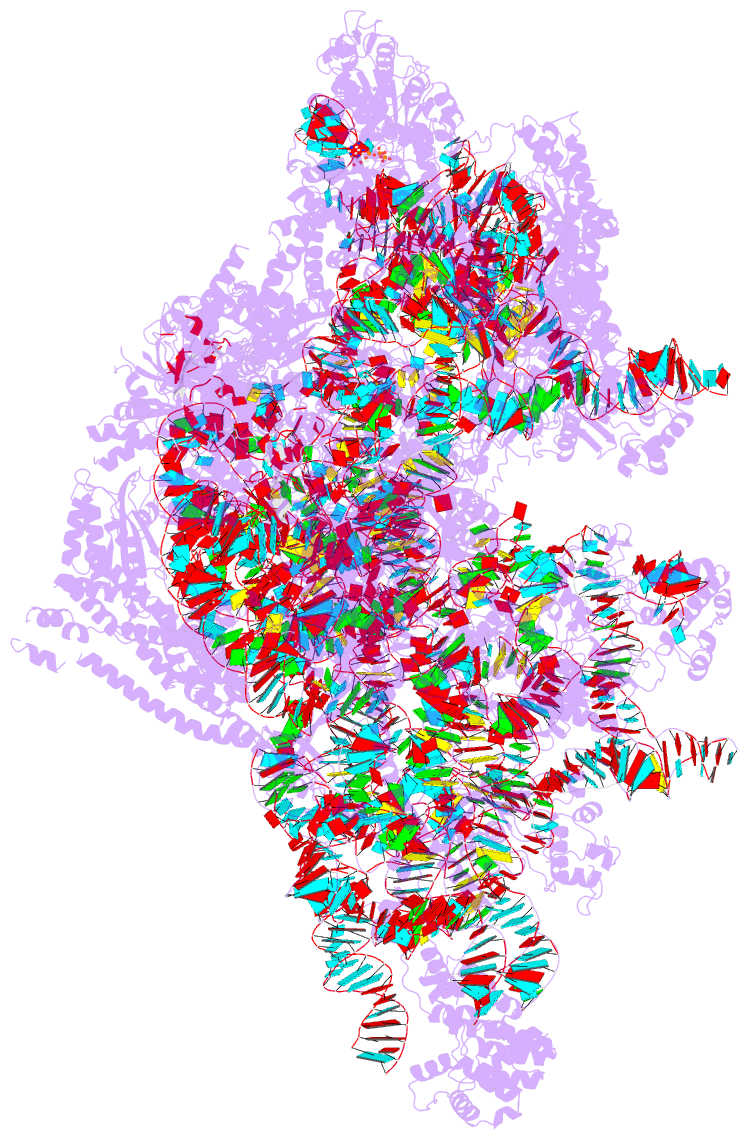
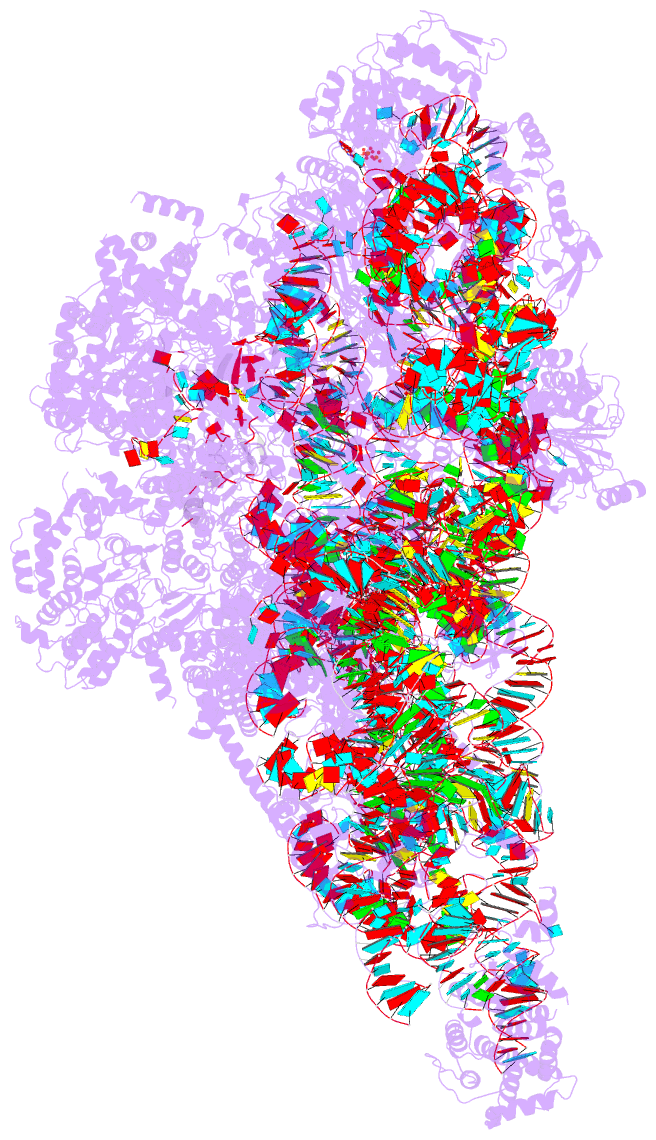
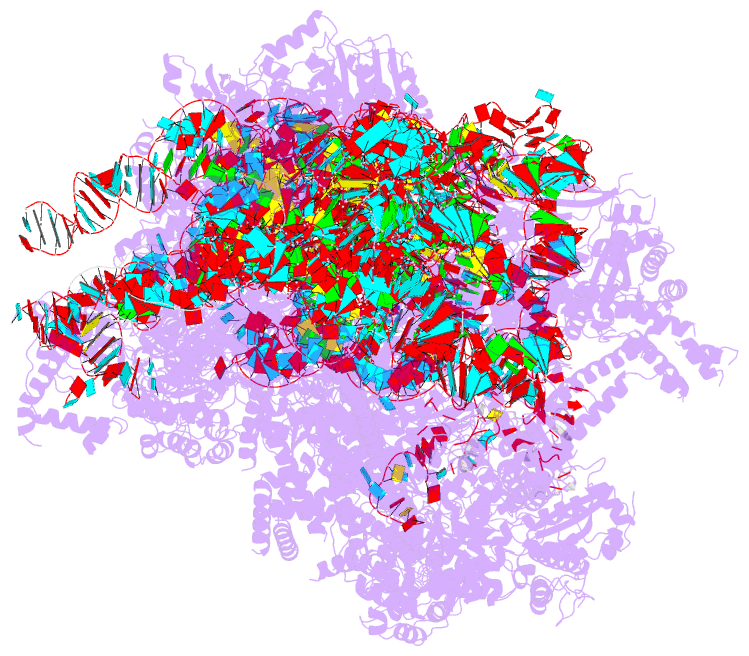
Summary information and primary citation
- PDB-id
- 8d8l; DSSR-derived features in text and JSON formats
- Class
- ribosome
- Method
- cryo-EM (2.6 Å)
- Summary
- Yeast mitochondrial small subunit assembly intermediate (state 3)
- Reference
- Harper NJ, Burnside C, Klinge S (2023): "Principles of mitoribosomal small subunit assembly in eukaryotes." Nature, 614, 175-181. doi: 10.1038/s41586-022-05621-0.
- Abstract
- Mitochondrial ribosomes (mitoribosomes) synthesize proteins encoded within the mitochondrial genome that are assembled into oxidative phosphorylation complexes. Thus, mitoribosome biogenesis is essential for ATP production and cellular metabolism1. Here we used cryo-electron microscopy to determine 9 structures of native yeast and human mitoribosomal small subunit assembly intermediates at resolutions from 2.4 to 3.8 Å, illuminating the mechanistic basis for how GTPases are employed to control early steps of decoding center formation, how initial rRNA folding and processing events are mediated, and how mitoribosomal proteins play active roles during assembly. Furthermore, this series of intermediates from two species with divergent mitoribosomal architecture uncovers both conserved principles and species-specific adaptations that govern the maturation of mitoribosomal small subunits in eukaryotes. By revealing the dynamic interplay between assembly factors, mitoribosomal proteins, and rRNA required to generate functional subunits, our structural analysis provides a vignette for how molecular complexity and diversity can evolve in large ribonucleoprotein assemblies.