Summary information and primary citation
- PDB-id
-
7syo;
SNAP-derived features in text and
JSON formats
- Class
- ribosome
- Method
- cryo-EM (4.6 Å)
- Summary
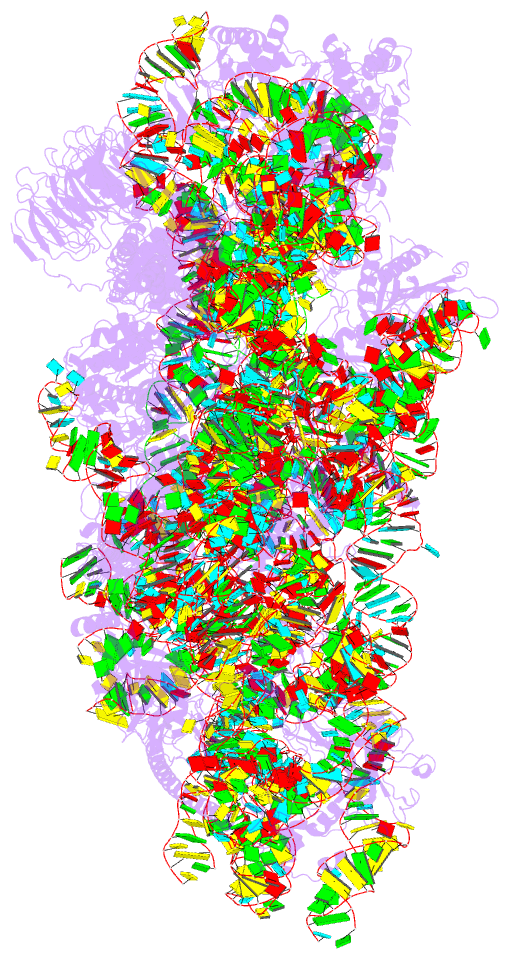
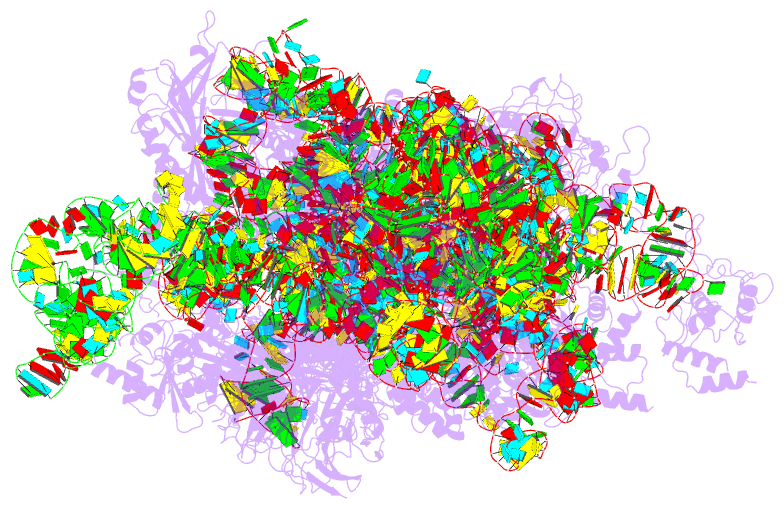
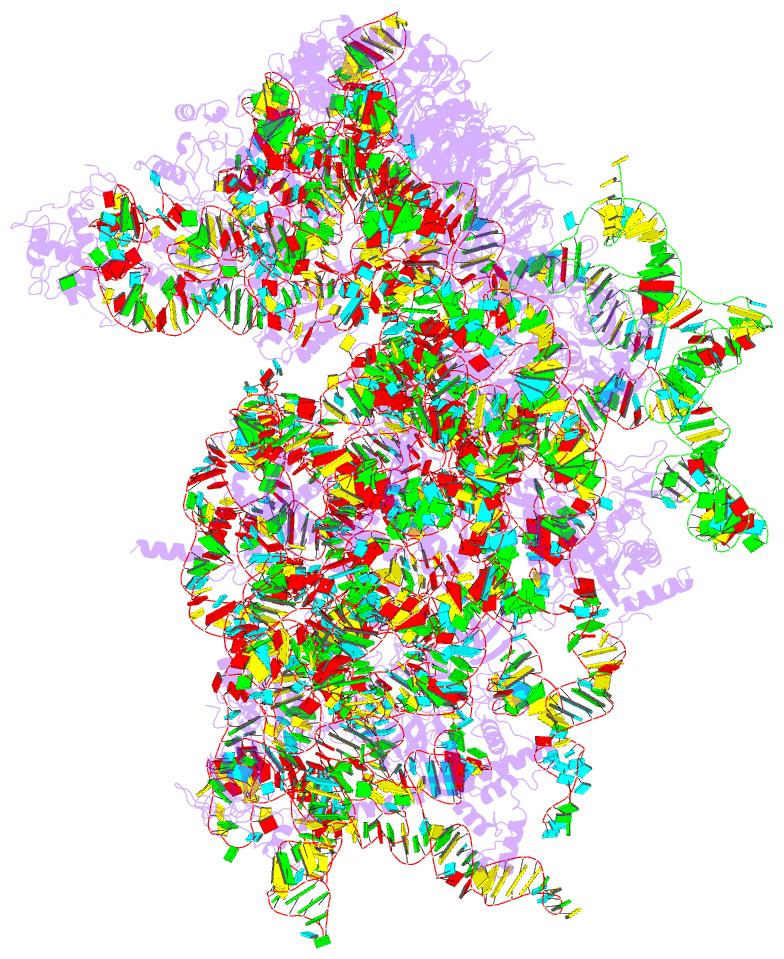
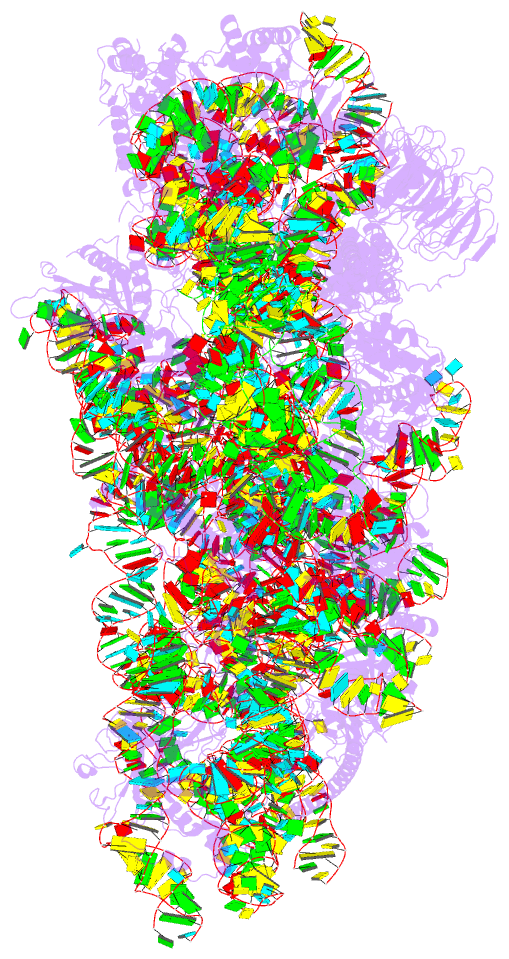
- Structure of the hcv ires bound to the 40s ribosomal
subunit, head open. structure 9(delta dii)
- Reference
-
Brown ZP, Abaeva IS, De S, Hellen CUT, Pestova TV, Frank
J (2022): "Molecular
architecture of 40S translation initiation complexes on
the hepatitis C virus IRES." Embo J.,
41, e110581. doi: 10.15252/embj.2022110581.
- Abstract
- Hepatitis C virus mRNA contains an internal ribosome
entry site (IRES) that mediates end-independent translation
initiation, requiring a subset of eukaryotic initiation
factors (eIFs). Biochemical studies revealed that direct
binding of the IRES to the 40S ribosomal subunit places the
initiation codon into the P site, where it base pairs with
eIF2-bound Met-tRNAiMet forming a 48S initiation complex.
Subsequently, eIF5 and eIF5B mediate subunit joining,
yielding an elongation-competent 80S ribosome. Initiation
can also proceed without eIF2, in which case Met-tRNAiMet
is recruited directly by eIF5B. However, the structures of
initiation complexes assembled on the HCV IRES, the
transitions between different states, and the accompanying
conformational changes have remained unknown. To fill these
gaps, we now obtained cryo-EM structures of IRES initiation
complexes, at resolutions up to 3.5 Å, that cover all major
stages from the initial ribosomal association, through
eIF2-containing 48S initiation complexes, to
eIF5B-containing complexes immediately prior to subunit
joining. These structures provide insights into the dynamic
network of 40S/IRES contacts, highlight the role of IRES
domain II, and reveal conformational changes that occur
during the transition from eIF2- to eIF5B-containing 48S
complexes and prepare them for subunit joining.