Summary information and primary citation
- PDB-id
-
7l4h;
SNAP-derived features in text and
JSON formats
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.56 Å)
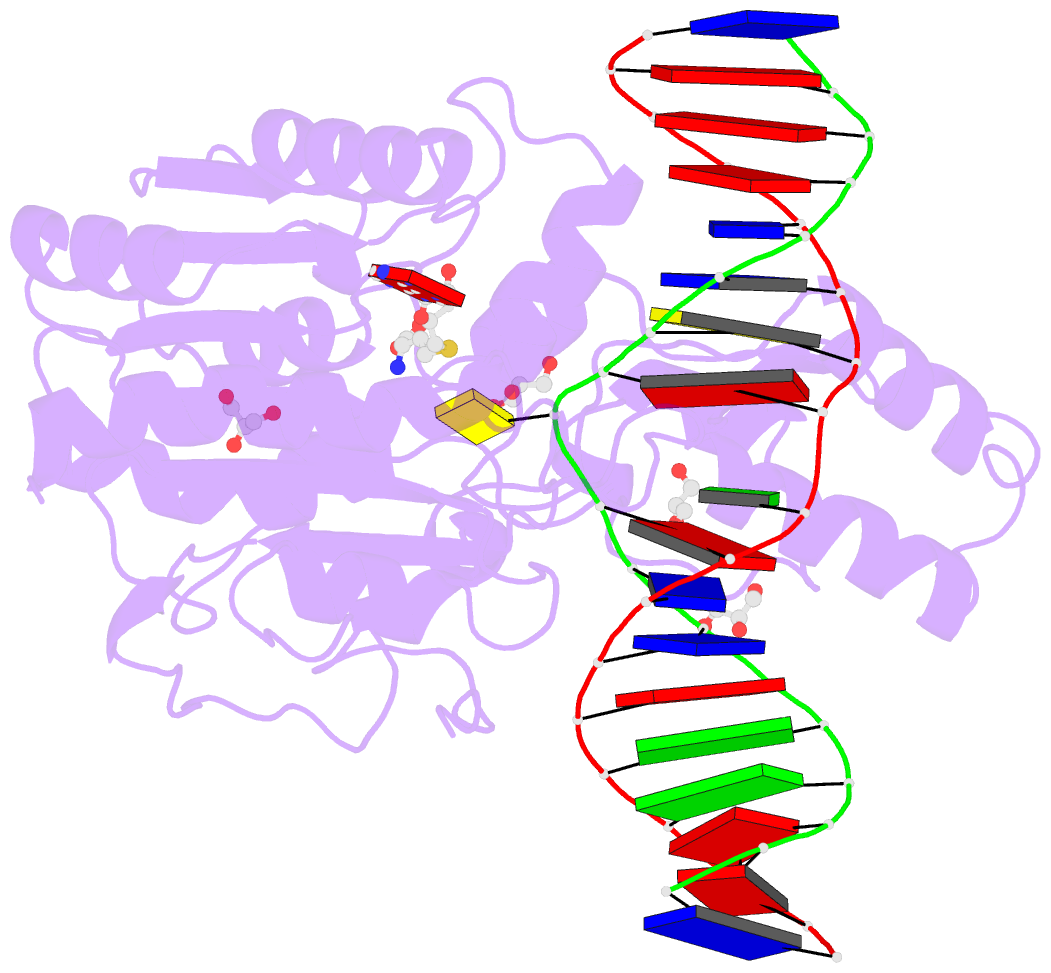
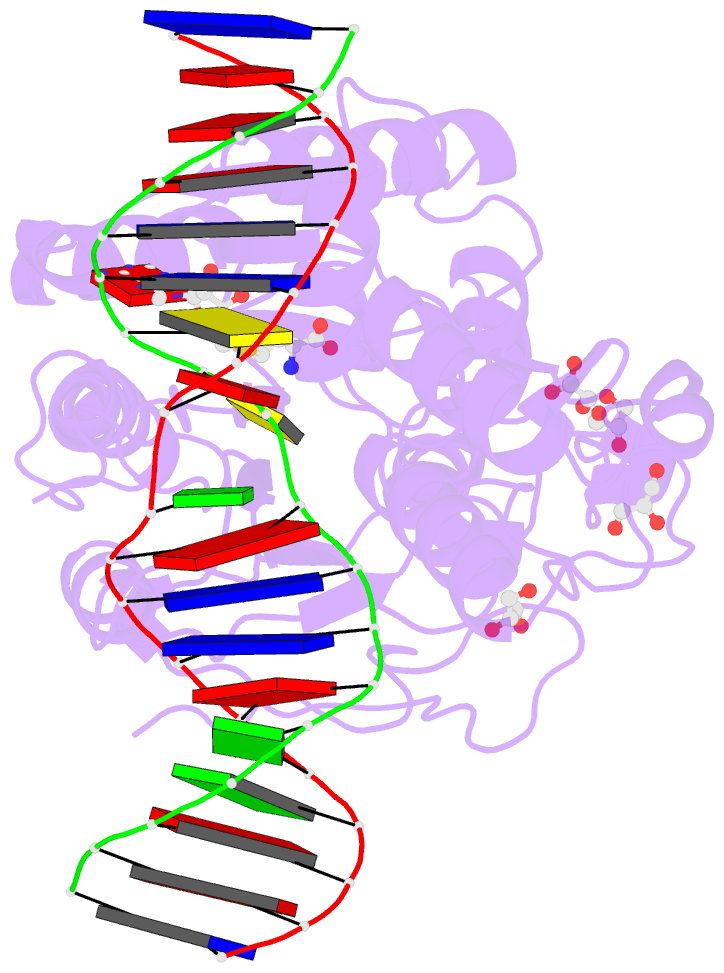
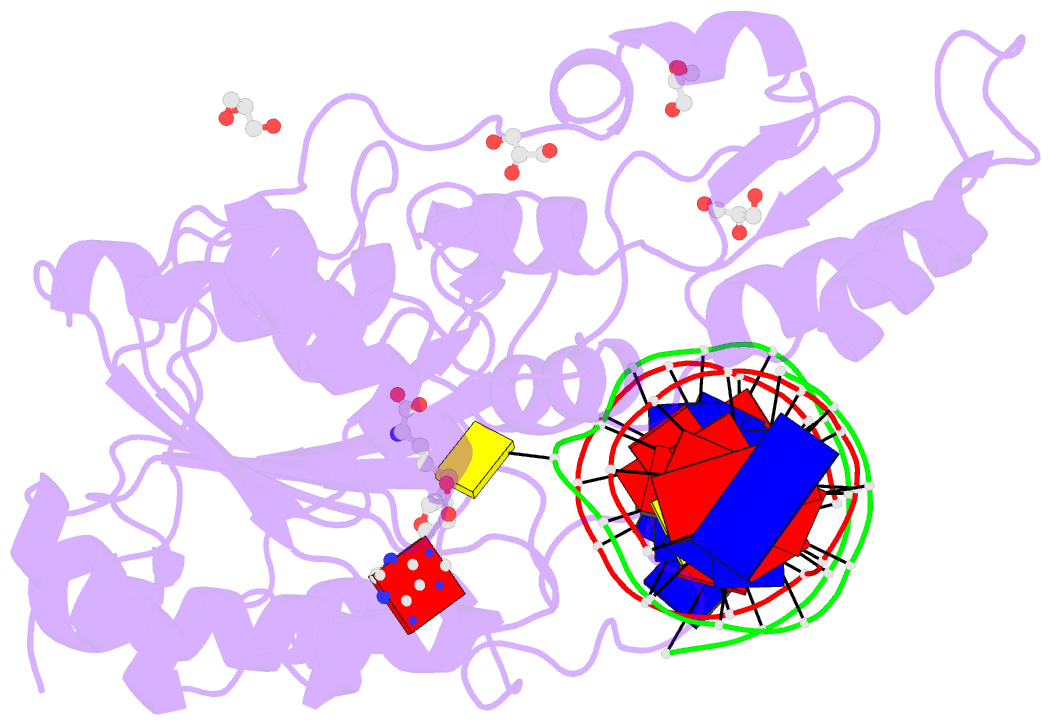
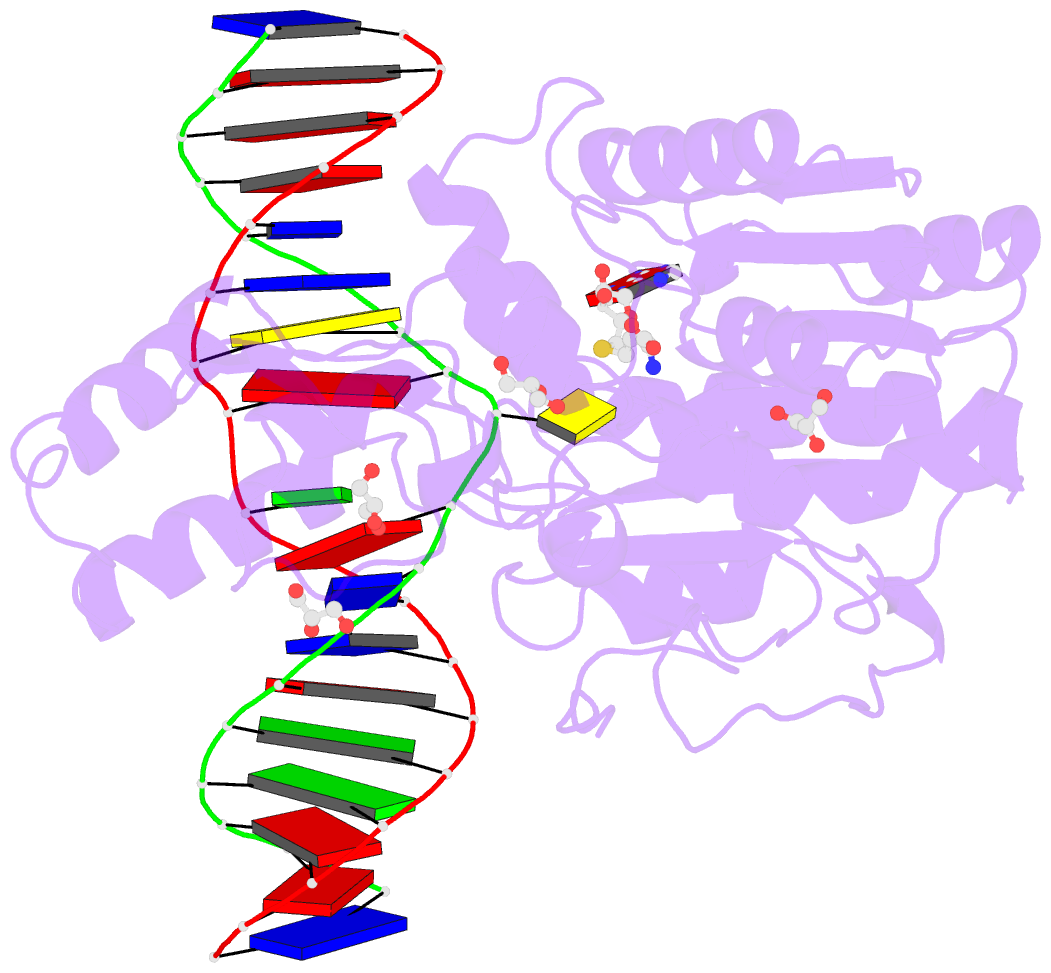
- Summary
- Crystal structure of the drm2-ctg DNA complex
- Reference
-
Fang J, Leichter SM, Jiang J, Biswal M, Lu J, Zhang ZM,
Ren W, Zhai J, Cui Q, Zhong X, Song J (2021): "Substrate
deformation regulates DRM2-mediated DNA methylation in
plants." Sci Adv, 7. doi:
10.1126/sciadv.abd9224.
- Abstract
- DNA methylation is a major epigenetic mechanism
critical for gene expression and genome stability. In
plants, domains rearranged methyltransferase 2 (DRM2)
preferentially mediates CHH (H = C, T, or A) methylation, a
substrate specificity distinct from that of mammalian DNA
methyltransferases. However, the underlying mechanism is
unknown. Here, we report structure-function
characterization of DRM2-mediated methylation. An arginine
finger from the catalytic loop intercalates into the
nontarget strand of DNA through the minor groove, inducing
large DNA deformation that affects the substrate preference
of DRM2. The target recognition domain stabilizes the
enlarged major groove via shape complementarity rather than
base-specific interactions, permitting substrate diversity.
The engineered DRM2 C397R mutation introduces base-specific
contacts with the +2-flanking guanine, thereby shifting the
substrate specificity of DRM2 toward CHG DNA. Together,
this study uncovers DNA deformation as a mechanism in
regulating the specificity of DRM2 toward diverse CHH
substrates and illustrates methylome complexity in
plants.