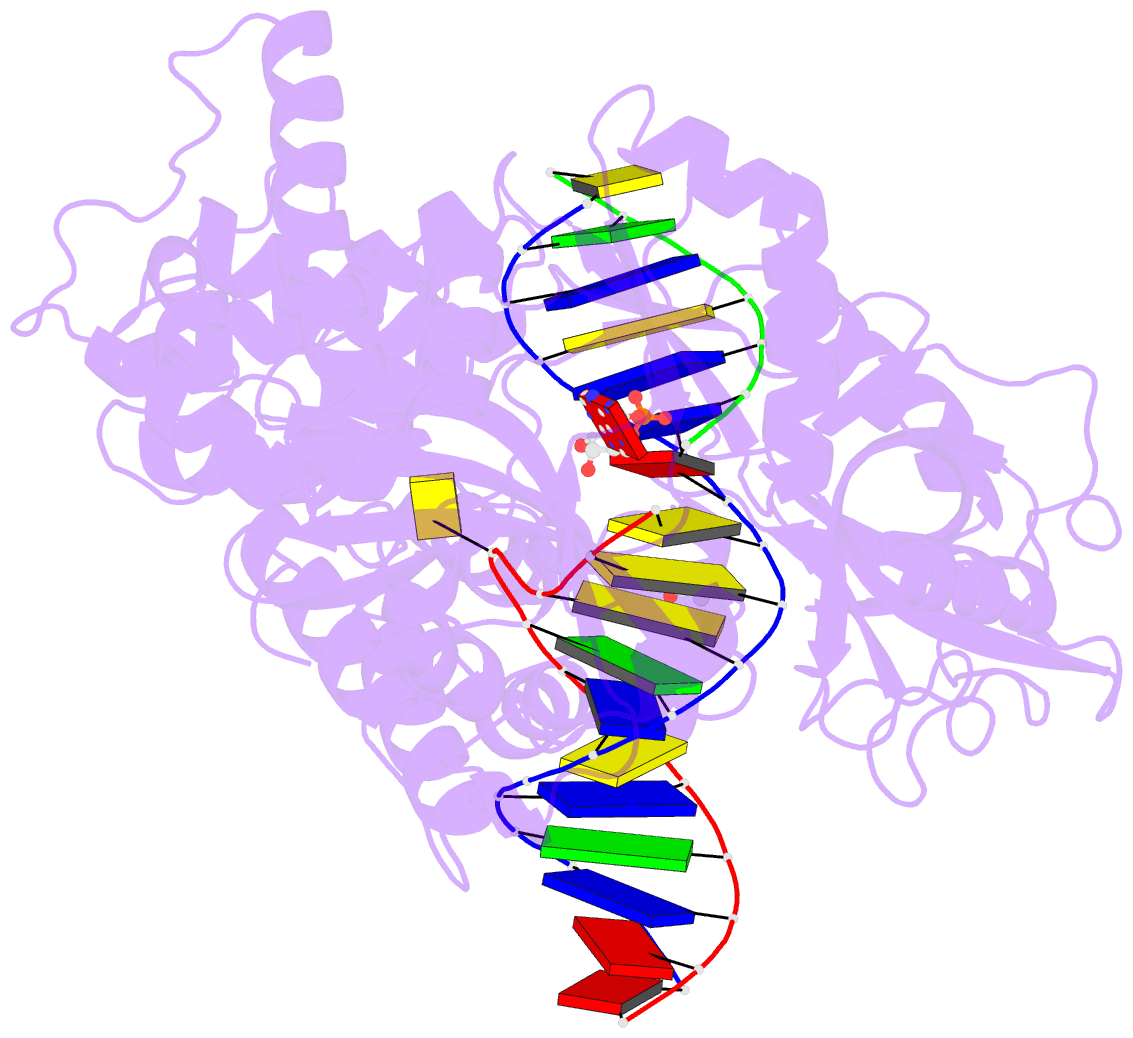
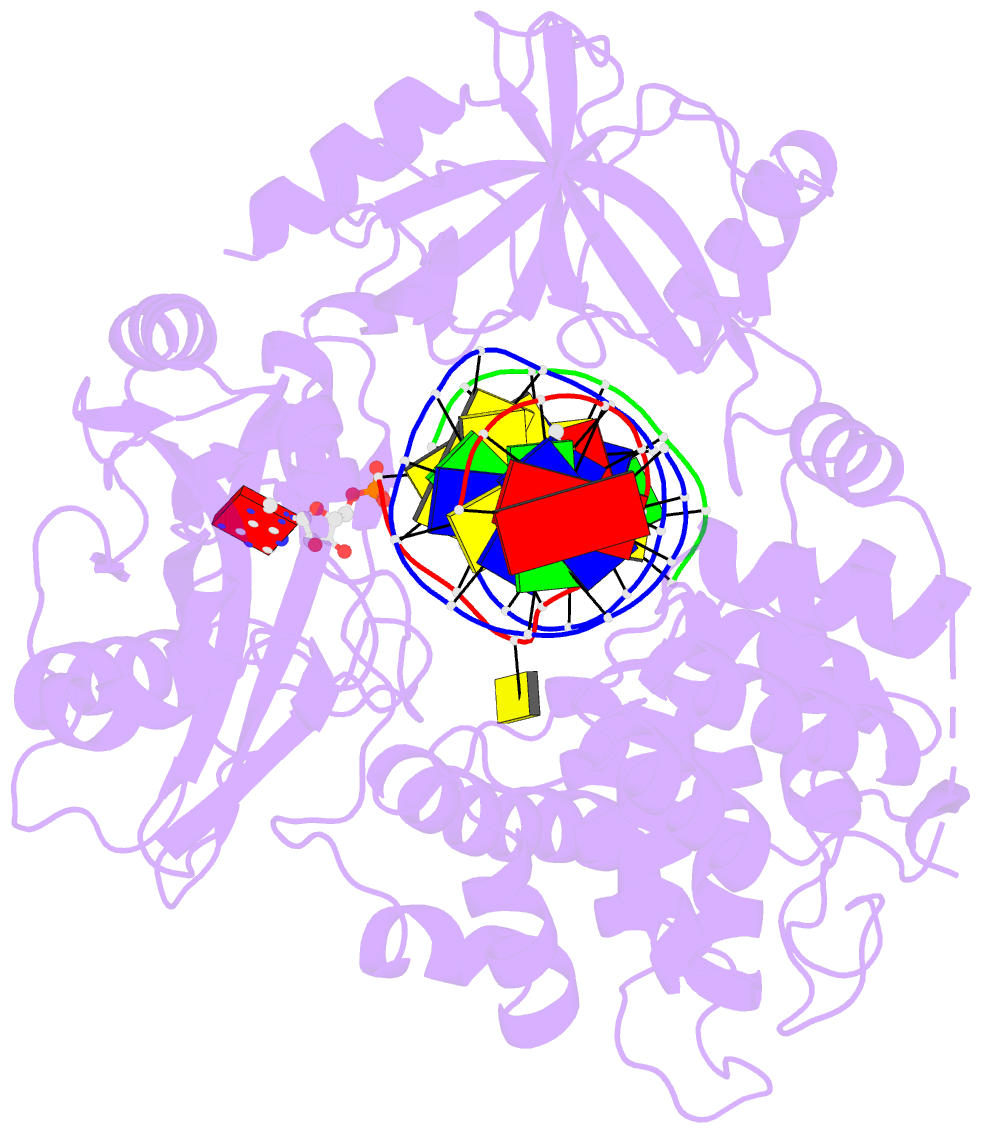
Summary information and primary citation
- PDB-id
-
7kr3;
SNAP-derived features in text and
JSON formats
- Class
- ligase-DNA
- Method
- X-ray (2.778 Å)
- Summary
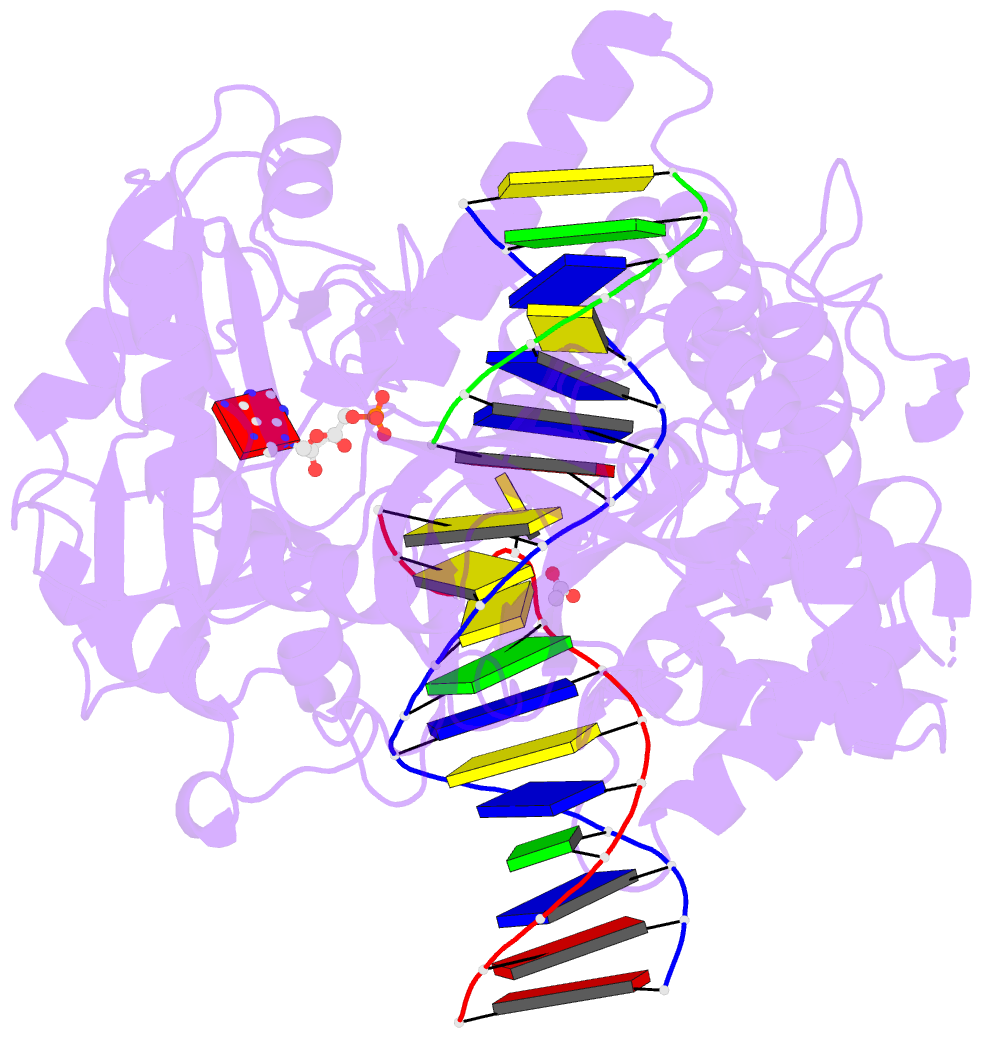
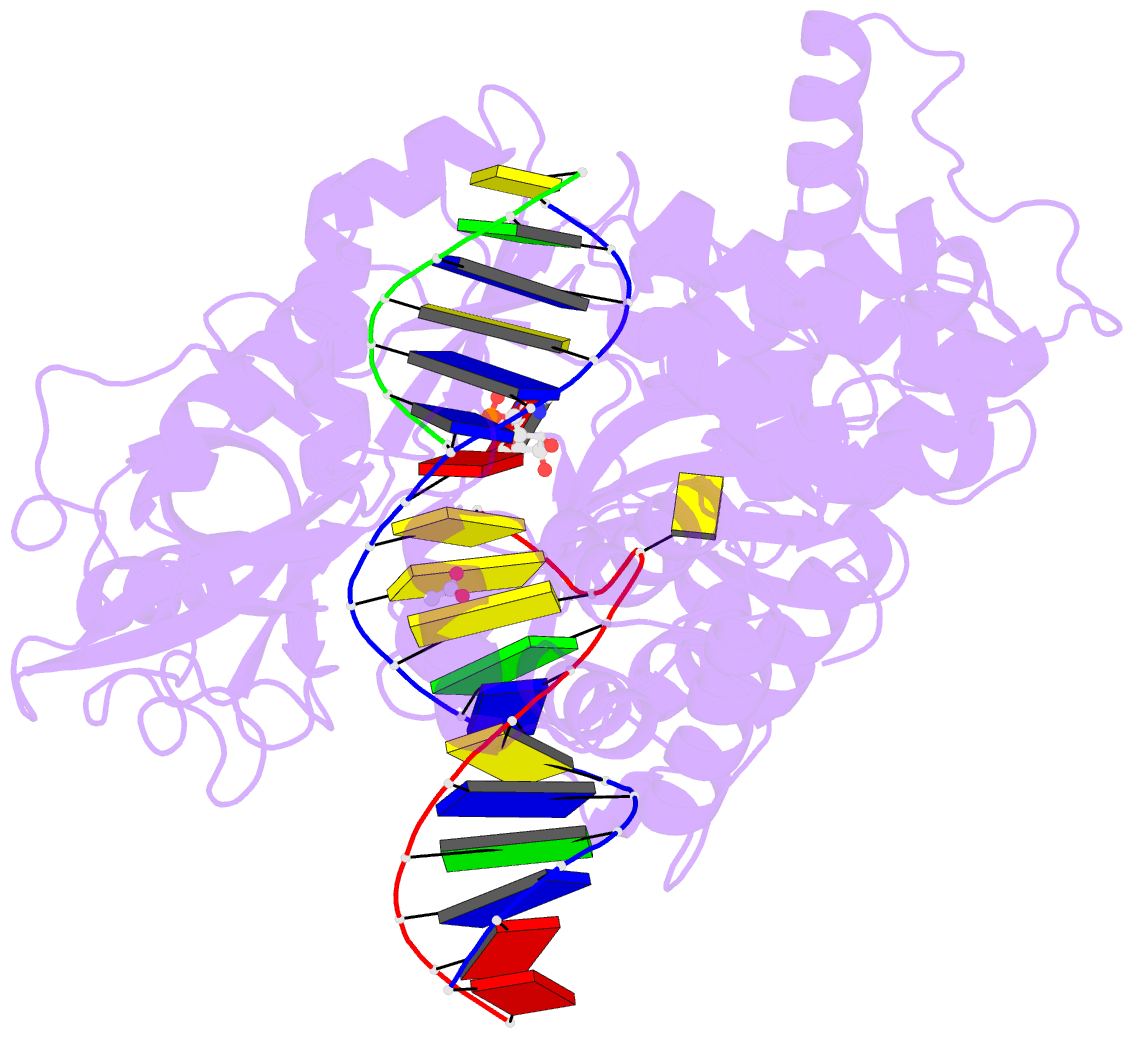
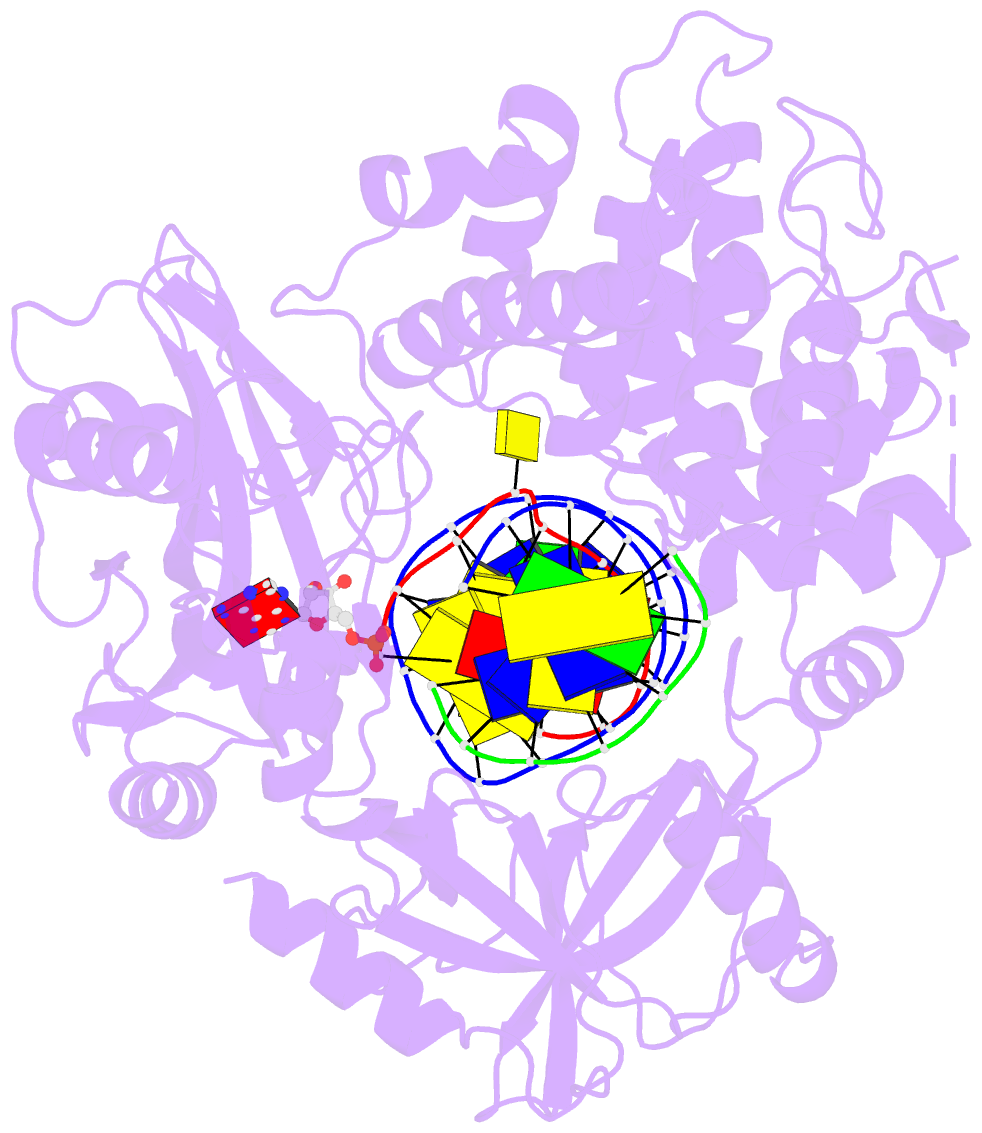
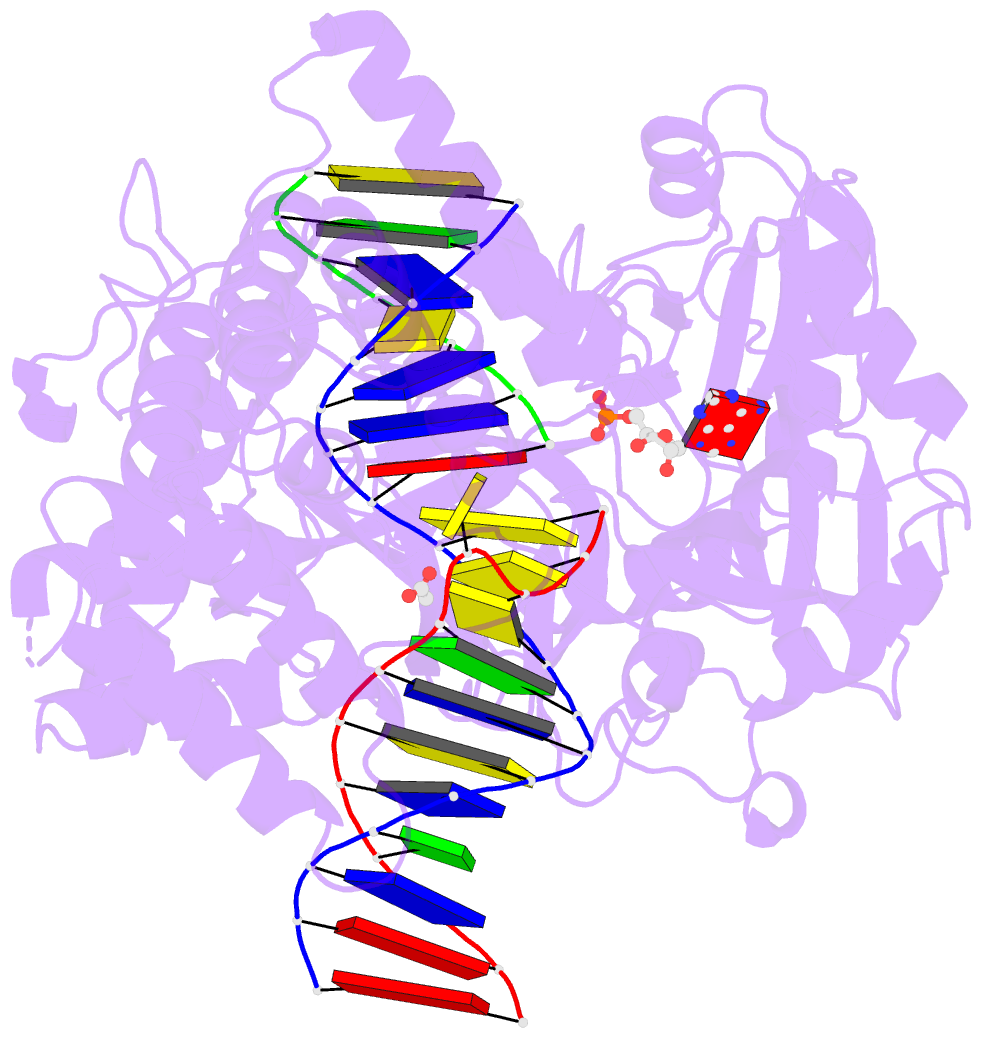
- Human DNA ligase 1(e346a-e592a) bound to a bulged DNA
substrate
- Reference
-
Williams JS, Tumbale PP, Arana ME, Rana JA, Williams RS,
Kunkel TA (2021): "High-fidelity
DNA ligation enforces accurate Okazaki fragment
maturation during DNA replication." Nat
Commun, 12, 482. doi: 10.1038/s41467-020-20800-1.
- Abstract
- DNA ligase 1 (LIG1, Cdc9 in yeast) finalizes eukaryotic
nuclear DNA replication by sealing Okazaki fragments using
DNA end-joining reactions that strongly discriminate
against incorrectly paired DNA substrates. Whether
intrinsic ligation fidelity contributes to the accuracy of
replication of the nuclear genome is unknown. Here, we show
that an engineered low-fidelity
LIG1<sub>Cdc9</sub> variant confers a novel
mutator phenotype in yeast typified by the accumulation of
single base insertion mutations in homonucleotide runs. The
rate at which these additions are generated increases upon
concomitant inactivation of DNA mismatch repair, or by
inactivation of the Fen1<sub>Rad27</sub>
Okazaki fragment maturation (OFM) nuclease. Biochemical and
structural data establish that
LIG1<sub>Cdc9</sub> normally avoids erroneous
ligation of DNA polymerase slippage products, and this
protection is compromised by mutation of a
LIG1<sub>Cdc9</sub> high-fidelity metal binding
site. Collectively, our data indicate that high-fidelity
DNA ligation is required to prevent insertion mutations,
and that this may be particularly critical following strand
displacement synthesis during the completion of OFM.