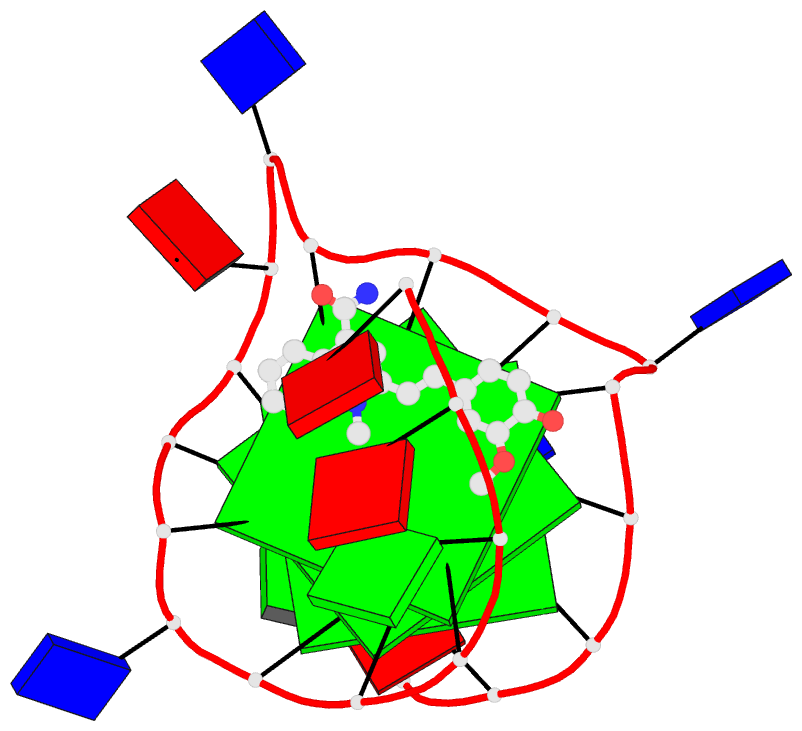
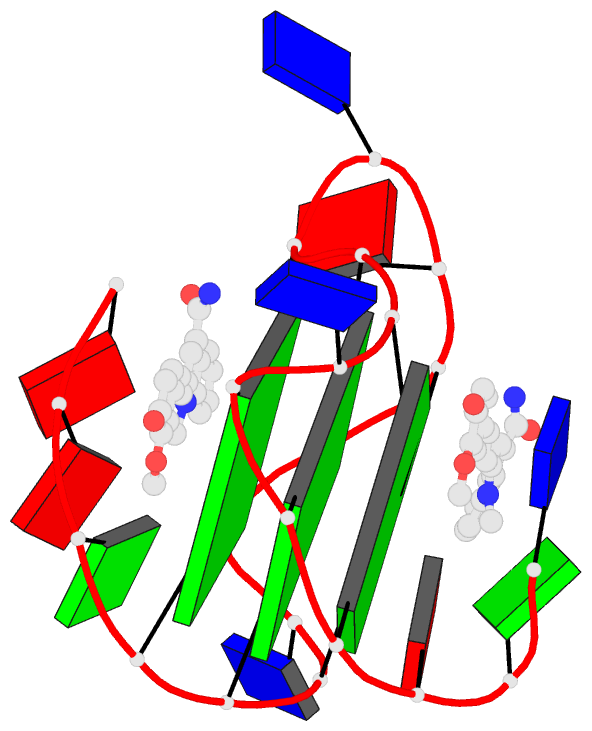
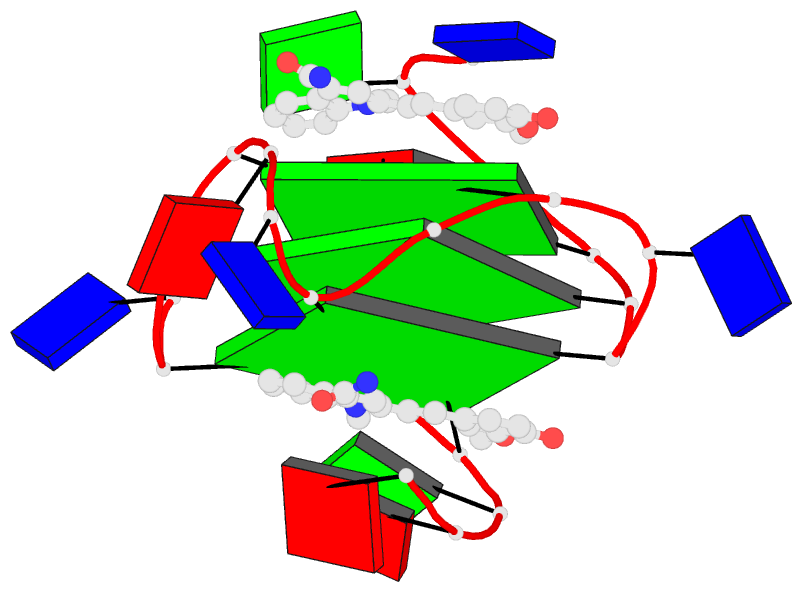
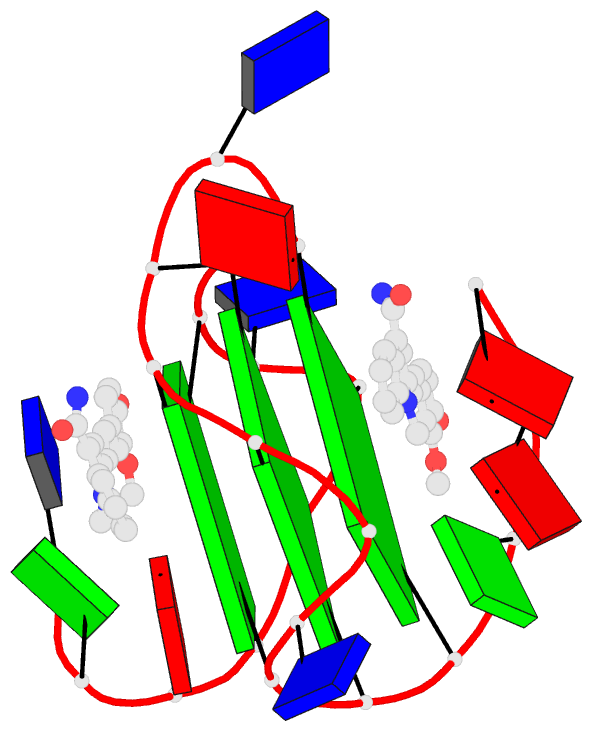
Summary information and primary citation
- PDB-id
-
7kbw;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Solution structure of the major myc promoter
g-quadruplex with a wild-type flanking in complex with
nsc85697, a quinoline derivative
- Reference
-
Dickerhoff J, Dai J, Yang D (2021): "Structural
recognition of the MYC promoter G-quadruplex by a
quinoline derivative: insights into molecular targeting
of parallel G-quadruplexes." Nucleic Acids
Res., 49, 5905-5915. doi: 10.1093/nar/gkab330.
- Abstract
- DNA G-Quadruplexes (G4s) formed in oncogene promoters
regulate transcription. The oncogene MYC promoter G4
(MycG4) is the most prevalent G4 in human cancers. However,
the most studied MycG4 sequence bears a mutated 3'-residue
crucial for ligand recognition. Here, we report a new
drug-like small molecule PEQ without a large aromatic
moiety that specifically binds MycG4. We determined the NMR
solution structures of the wild-type MycG4 and its 2:1 PEQ
complex, as well as the structure of the 2:1 PEQ complex of
the widely used mutant MycG4. Comparison of the two complex
structures demonstrates specific molecular recognition of
MycG4 and shows the clear effect of the critical
3'-mutation on the drug binding interface. We performed a
systematic analysis of the four available complex
structures involving the same mutant MycG4, which can be
considered a model system for parallel G4s, and revealed
for the first time that the flexible flanking residues are
recruited in a conserved and sequence-specific way, as well
as unused potential for selective ligand-G4 hydrogen-bond
interactions. Our results provide the true molecular basis
for MycG4-targeting drugs and new critical insights into
future rational design of drugs targeting MycG4 and
parallel G4s that are prevalent in promoter and RNA
G4s.