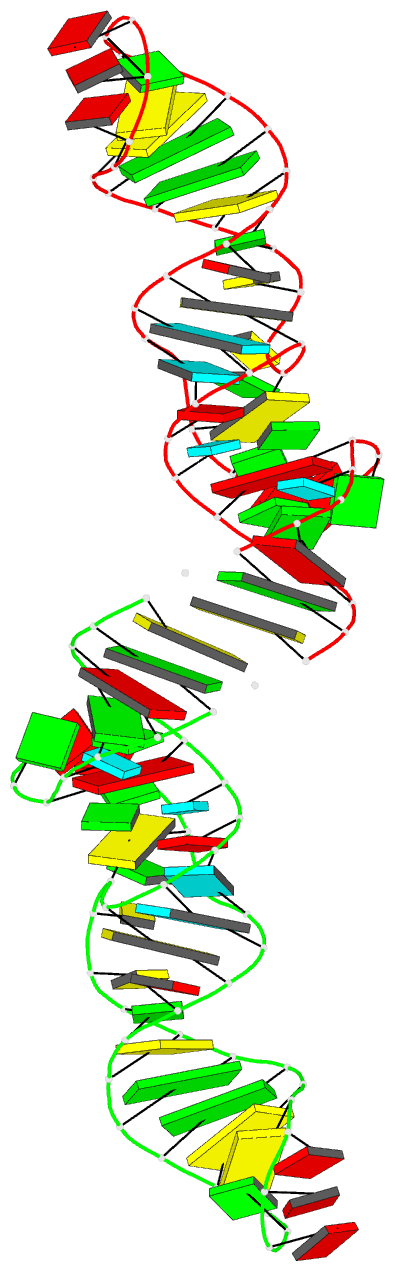
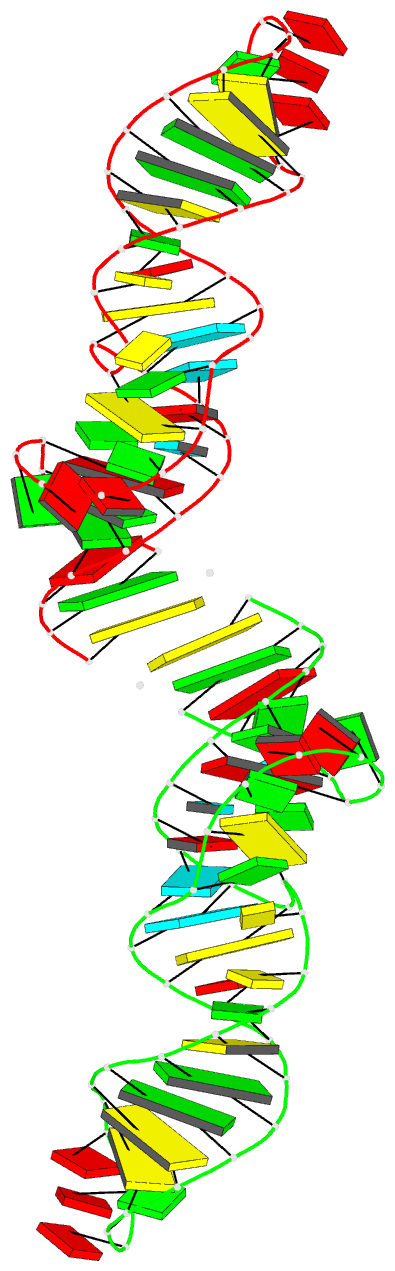
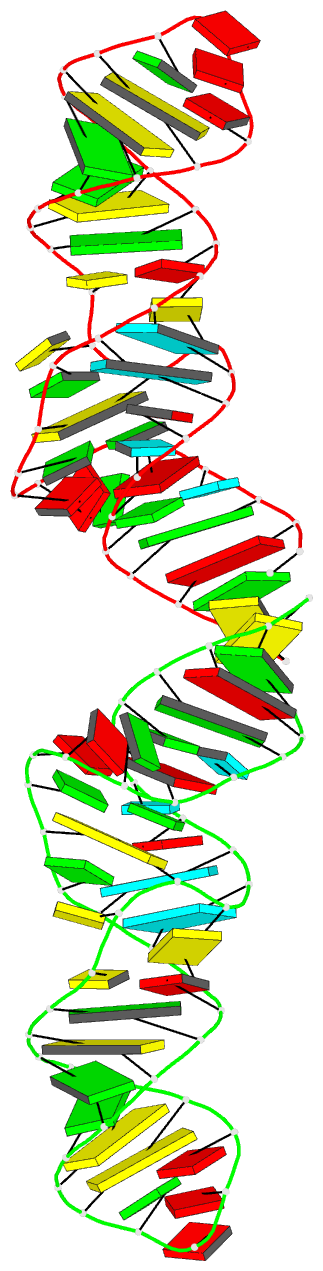
Summary information and primary citation
- PDB-id
-
7elp;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (2.79 Å)
- Summary
- Crystal structure of xanthine riboswitch with xanthine,
iridium hexammine soak
- Reference
-
Xu X, Egger M, Chen H, Bartosik K, Micura R, Ren A
(2021): "Insights
into xanthine riboswitch structure and metal ion-mediated
ligand recognition." Nucleic Acids Res.,
49, 7139-7153. doi: 10.1093/nar/gkab486.
- Abstract
- Riboswitches are conserved functional domains in mRNA
that mostly exist in bacteria. They regulate gene
expression in response to varying concentrations of
metabolites or metal ions. Recently, the NMT1 RNA motif has
been identified to selectively bind xanthine and uric acid,
respectively, both are involved in the metabolic pathway of
purine degradation. Here, we report a crystal structure of
this RNA bound to xanthine. Overall, the riboswitch
exhibits a rod-like, continuously stacked fold composed of
three stems and two internal junctions. The binding-pocket
is determined by the highly conserved junctional sequence
J1 between stem P1 and P2a, and engages a long-distance
Watson-Crick base pair to junction J2. Xanthine inserts
between a G-U pair from the major groove side and is
sandwiched between base triples. Strikingly, a Mg2+ ion is
inner-sphere coordinated to O6 of xanthine and a
non-bridging oxygen of a backbone phosphate. Two further
hydrated Mg2+ ions participate in extensive interactions
between xanthine and the pocket. Our structure model is
verified by ligand binding analysis to selected riboswitch
mutants using isothermal titration calorimetry, and by
fluorescence spectroscopic analysis of RNA folding using
2-aminopurine-modified variants. Together, our study
highlights the principles of metal ion-mediated ligand
recognition by the xanthine riboswitch.