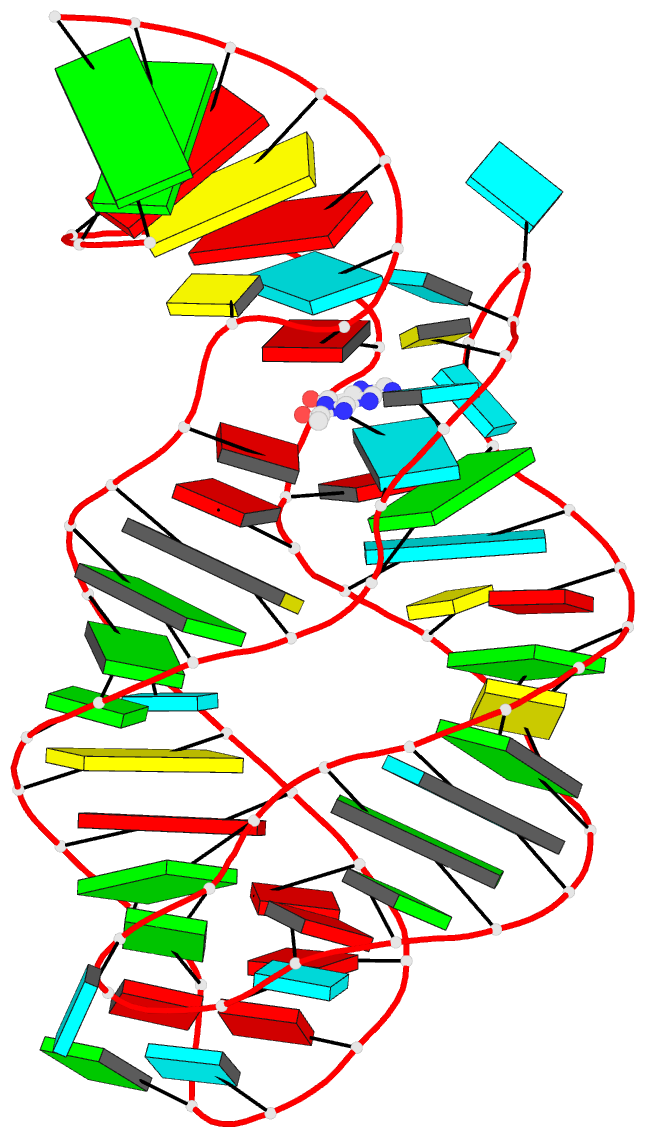
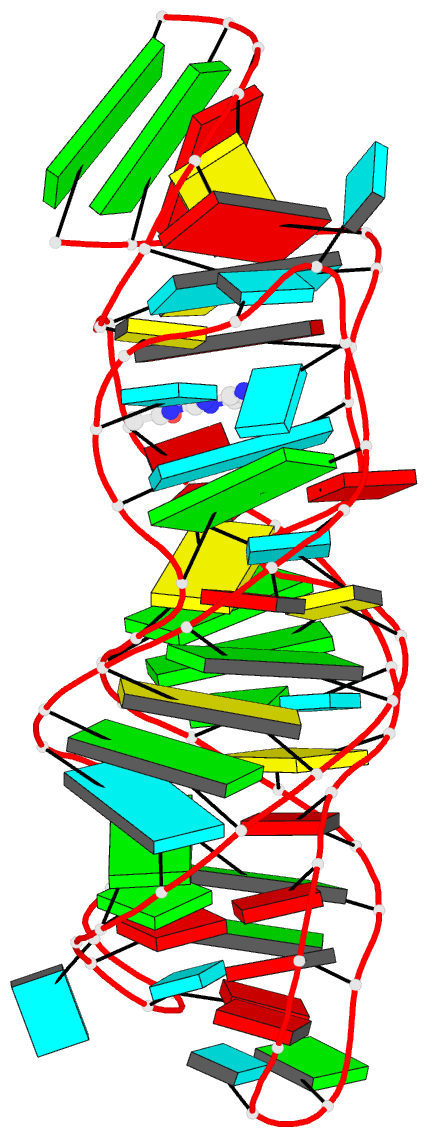
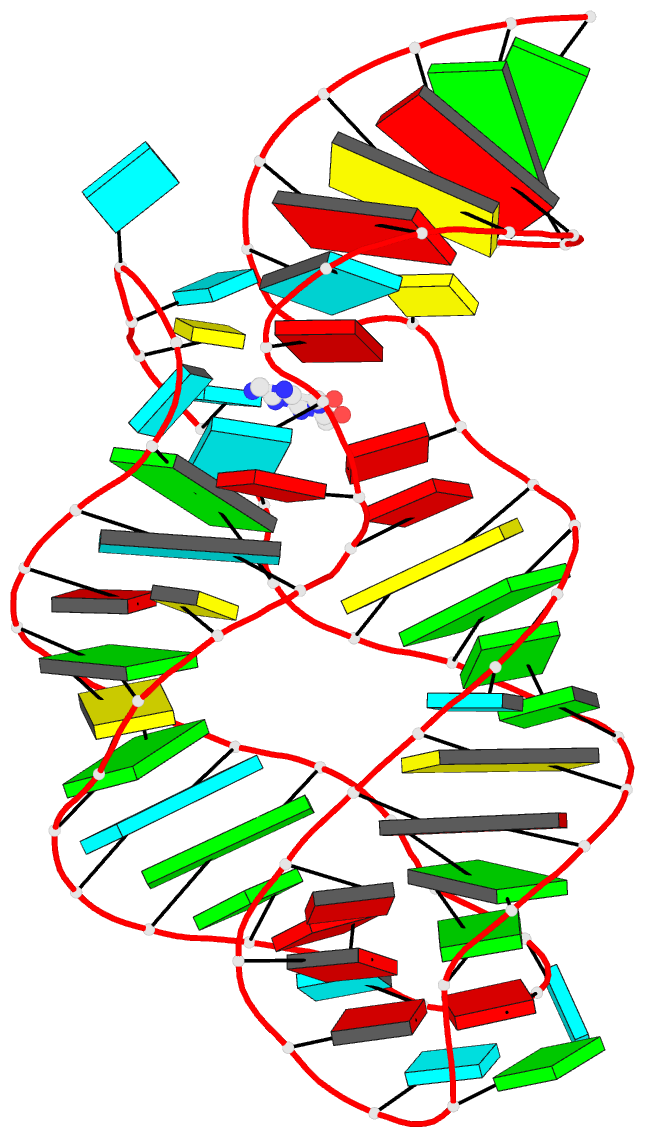
Summary information and primary citation
- PDB-id
-
6uc7;
SNAP-derived features in text and
JSON formats
- Class
- RNA
- Method
- X-ray (1.798 Å)
- Summary
- Structure of guanine riboswitch bound to n2-acetyl
guanine
- Reference
-
Matyjasik MM, Hall SD, Batey RT (2020): "High
Affinity Binding of N2-Modified Guanine Derivatives
Significantly Disrupts the Ligand Binding Pocket of the
Guanine Riboswitch." Molecules,
25. doi: 10.3390/molecules25102295.
- Abstract
- Riboswitches are important model systems for the
development of approaches to search for RNA-targeting
therapeutics. A principal challenge in finding compounds
that target riboswitches is that the effector ligand is
typically almost completely encapsulated by the RNA, which
severely limits the chemical space that can be explored.
Efforts to find compounds that bind the guanine/adenine
class of riboswitches with a high affinity have in part
focused on purines modified at the C6 and C2 positions.
These studies have revealed compounds that have low to
sub-micromolar affinity and, in a few cases, have
antimicrobial activity. To further understand how these
compounds interact with the guanine riboswitch, we have
performed an integrated structural and functional analysis
of representative guanine derivatives with modifications at
the C8, C6 and C2 positions. Our data indicate that while
modifications of guanine at the C6 position are generally
unfavorable, modifications at the C8 and C2 positions yield
compounds that rival guanine with respect to binding
affinity. Surprisingly, C2-modified guanines such as
<i>N</i>2-acetylguanine completely disrupt a
key Watson-Crick pairing interaction between the ligand and
RNA. These compounds, which also modulate transcriptional
termination as efficiently as guanine, open up a
significant new chemical space of guanine modifications in
the search for antimicrobial agents that target purine
riboswitches.