Summary information and primary citation
- PDB-id
-
5u01;
SNAP-derived features in text and
JSON formats
- Class
- transcription-DNA
- Method
- X-ray (2.5 Å)
- Summary
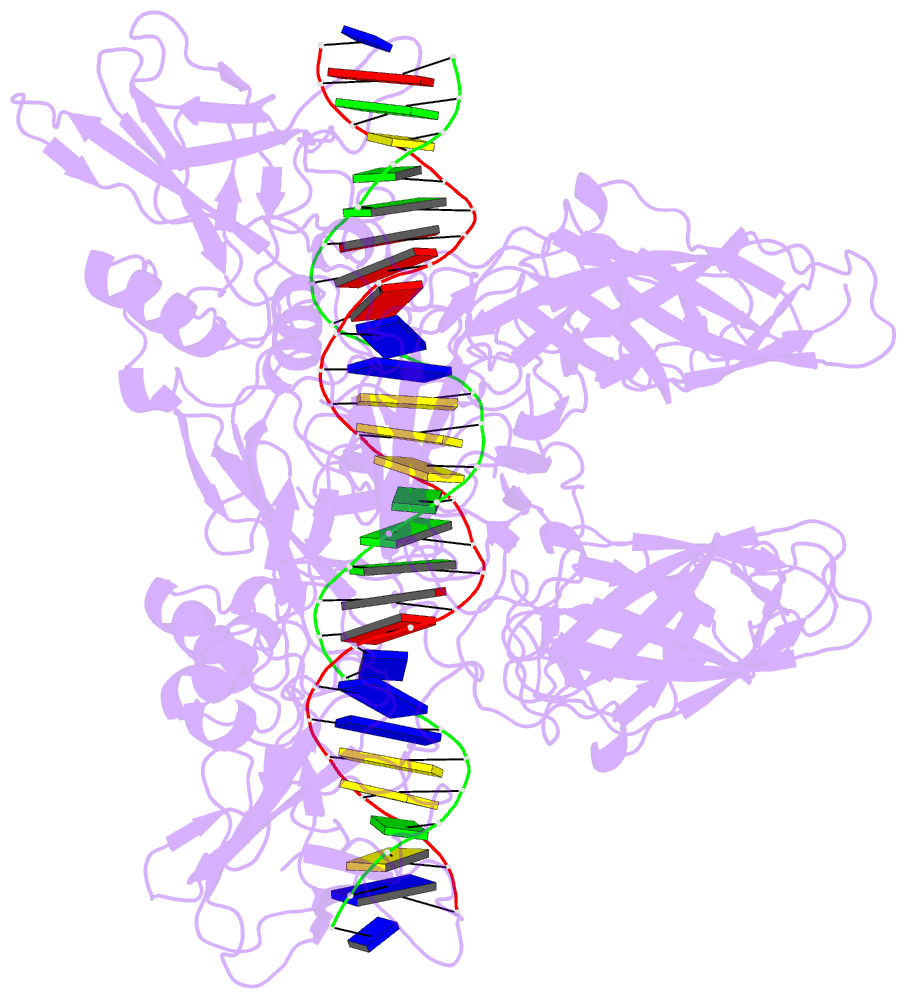
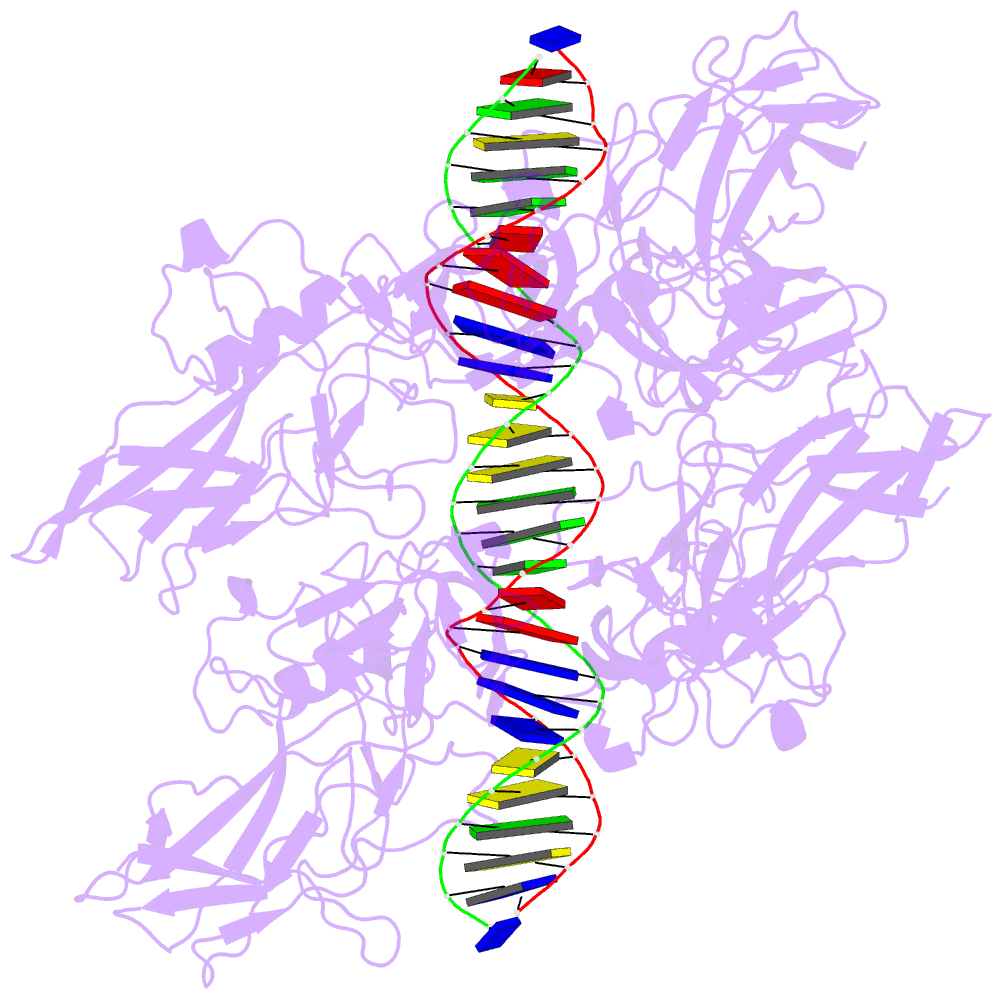
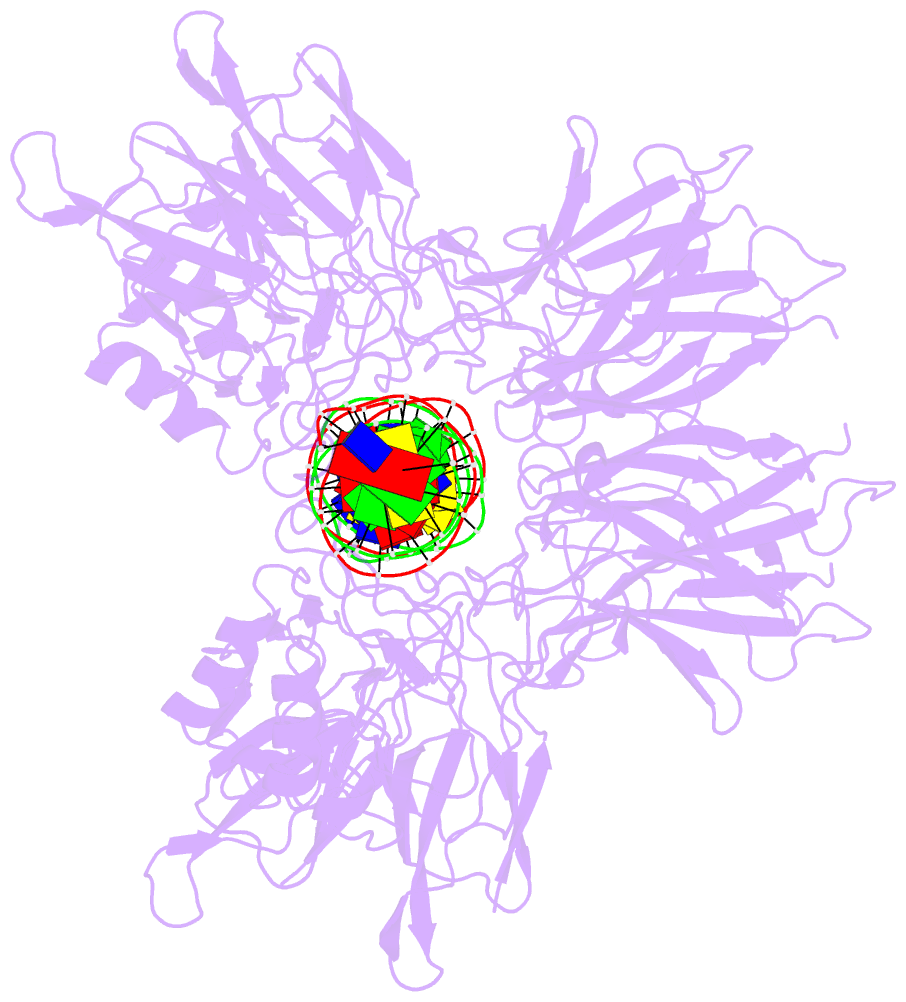
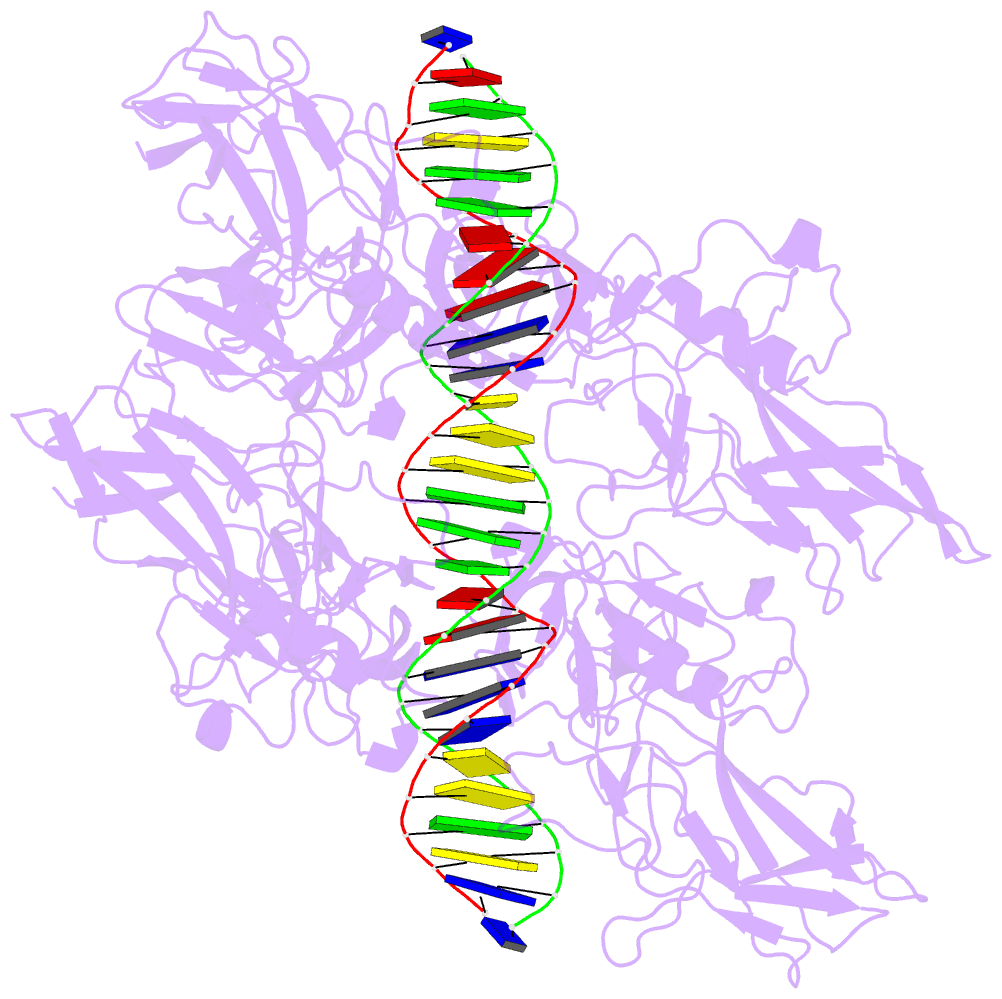
- Cooperative DNA binding by two rela dimers
- Reference
-
Mulero MC, Huang DB, Nguyen HT, Wang VY, Li Y, Biswas T,
Ghosh G (2017): "DNA-binding
affinity and transcriptional activity of the RelA
homodimer of nuclear factor kappa B are not
correlated." J. Biol. Chem.,
292, 18821-18830. doi: 10.1074/jbc.M117.813980.
- Abstract
- The nuclear factor κB (NF-κB) transcription factor
family regulates genes involved in cell proliferation and
inflammation. The promoters of these genes often contain
NF-κB-binding sites (κB sites) arranged in tandem. How
NF-κB activates transcription through these multiple sites
is incompletely understood. We report here an X-ray crystal
structure of homodimers comprising the RelA DNA-binding
domain containing the Rel homology region (RHR) in NF-κB
bound to an E-selectin promoter fragment with tandem κB
sites. This structure revealed that two dimers bind
asymmetrically to the symmetrically arranged κB sites at
which multiple cognate contacts between one dimer to the
corresponding DNA are broken. Because simultaneous RelA-RHR
dimer binding to tandem sites in solution was
anti-cooperative, we inferred that asymmetric RelA-RHR
binding with fewer contacts likely indicates a dissociative
binding mode. We found that both κB sites are essential for
reporter gene activation by full-length RelA homodimer,
suggesting that dimers facilitate DNA binding to each other
even though their stable co-occupation is not promoted.
Promoter variants with altered spacing and orientation of
tandem κB sites displayed unexpected reporter activities
that were not explained by the solution-binding pattern of
RelA-RHR. Remarkably, full-length RelA bound all DNAs with
a weaker affinity and specificity. Moreover, the
transactivation domain played a negative role in DNA
binding. These observations suggest that other nuclear
factors influence full-length RelA binding to DNA by
neutralizing the transactivation domain negative effect. We
propose that DNA binding by NF-κB dimers is highly complex
and modulated by facilitated association-dissociation
processes.