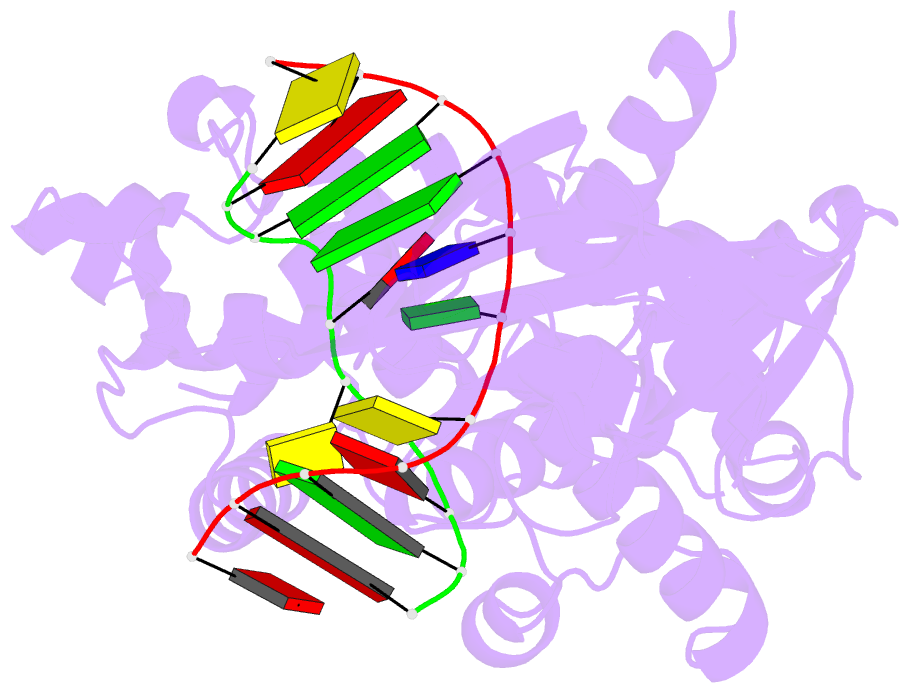
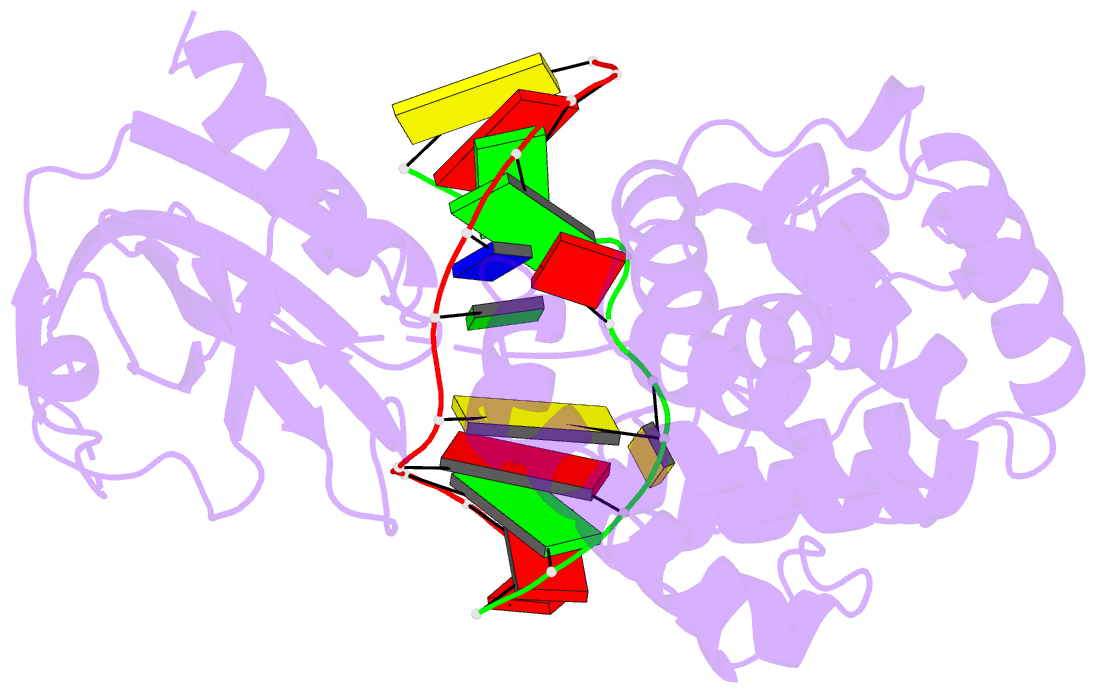
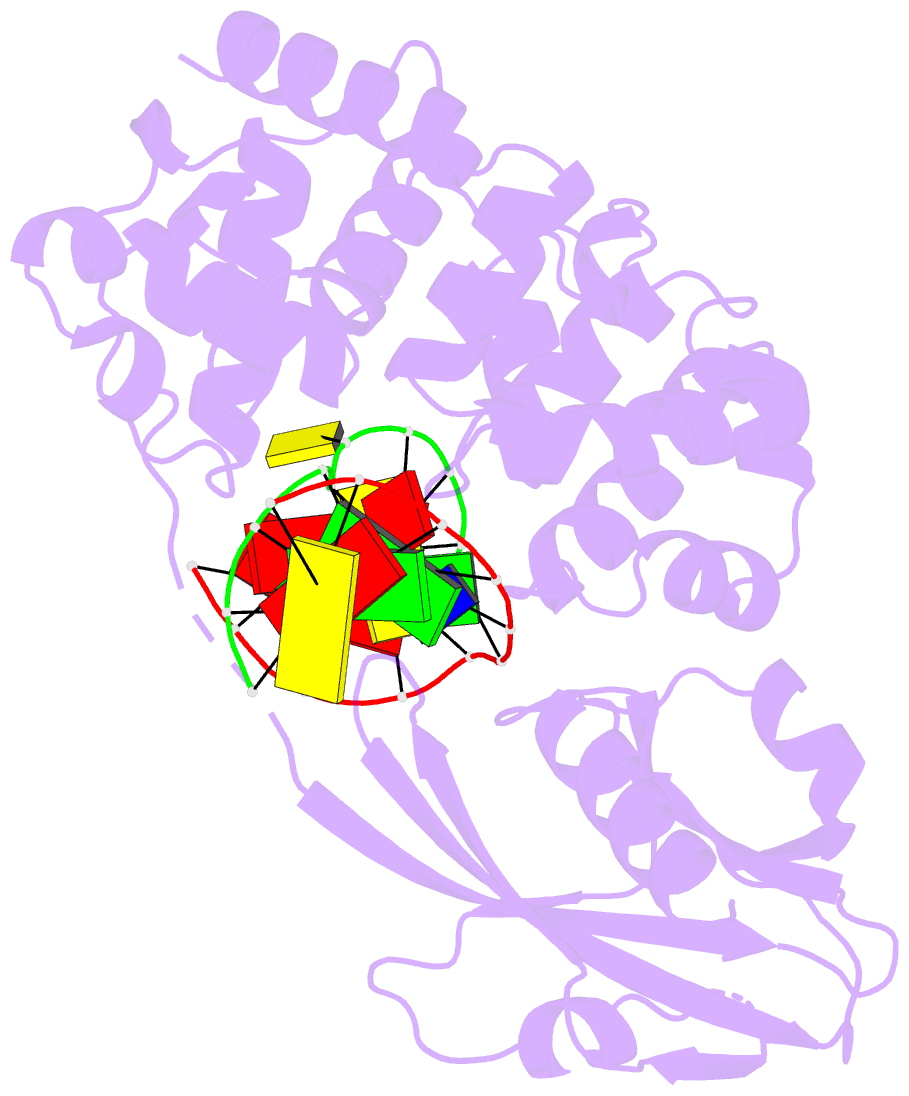
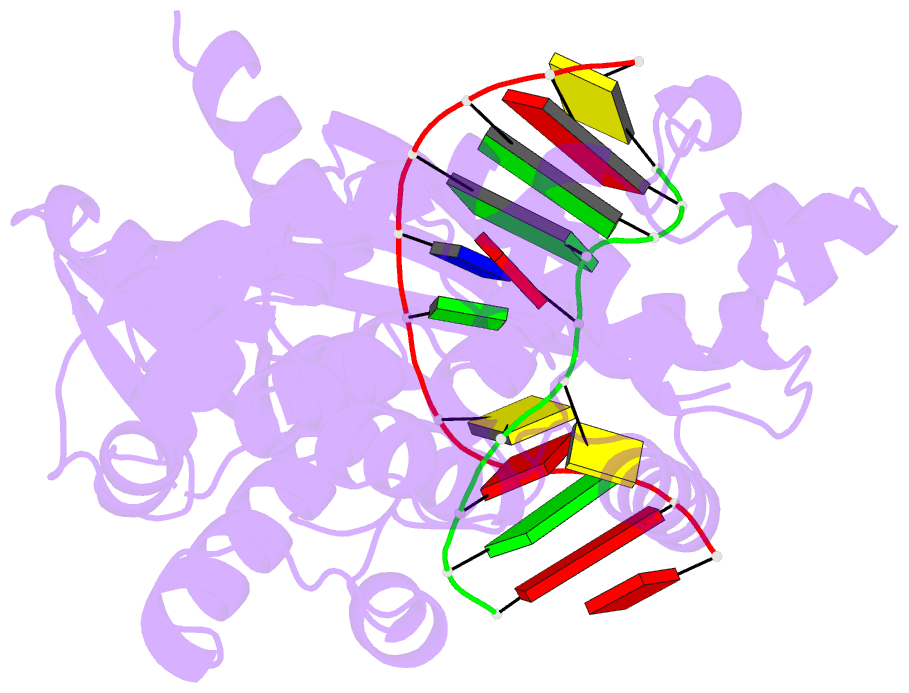
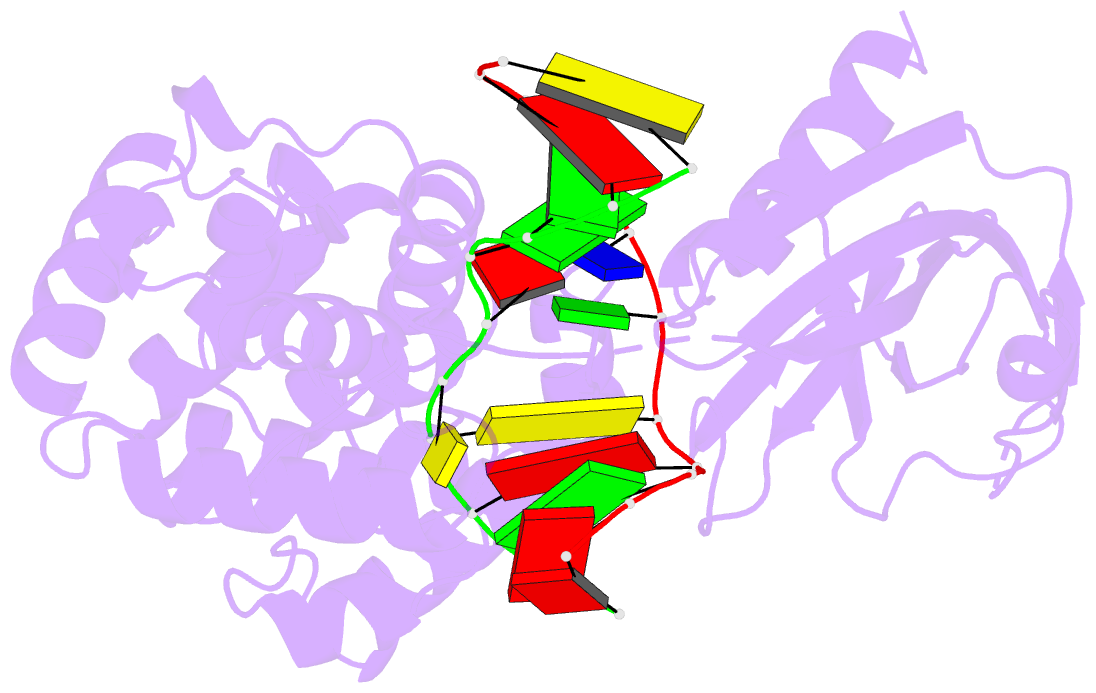
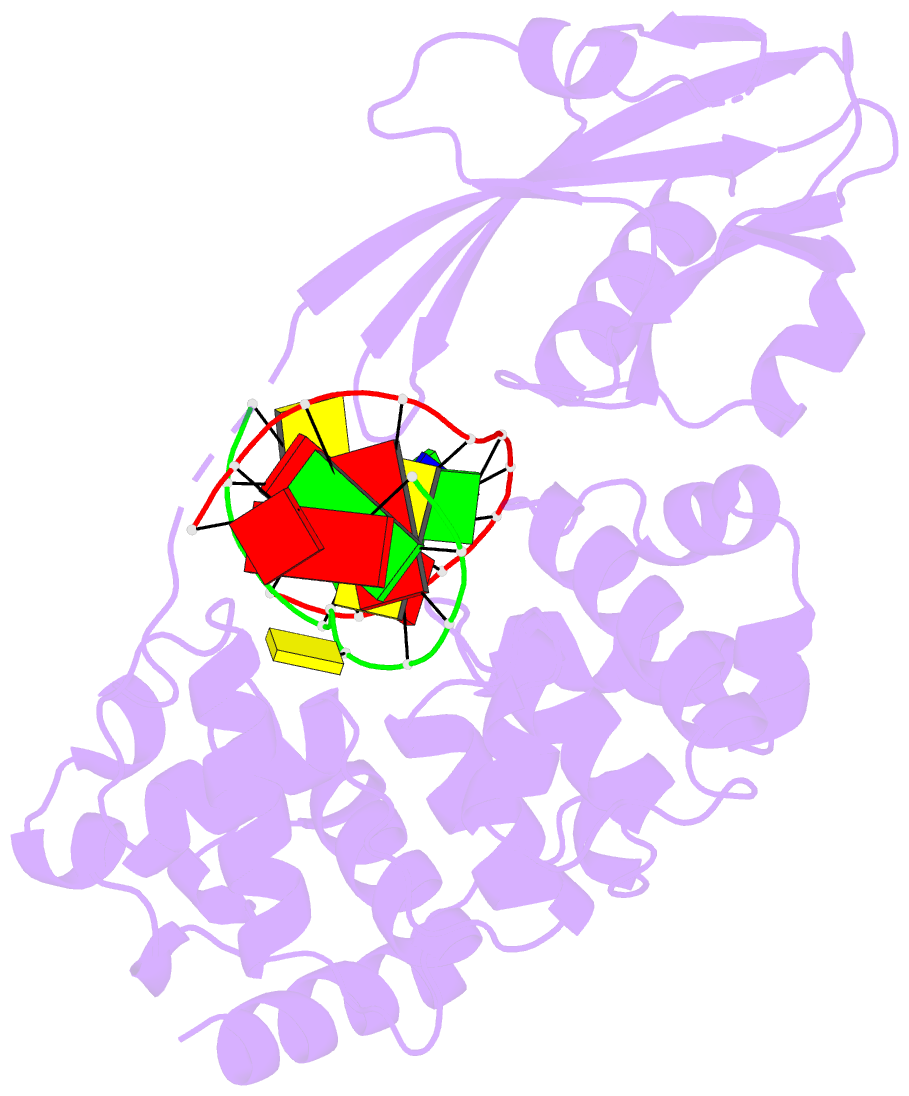
Summary information and primary citation
- PDB-id
-
4yph;
SNAP-derived features in text and
JSON formats
- Class
- hydrolase-DNA
- Method
- X-ray (2.32 Å)
- Summary
- Crystal structure of muty bound to its anti-substrate
with the disulfide cross-linker reduced
- Reference
-
Wang L, Lee SJ, Verdine GL (2015): "Structural
Basis for Avoidance of Promutagenic DNA Repair by MutY
Adenine DNA Glycosylase." J.Biol.Chem.,
290, 17096-17105. doi: 10.1074/jbc.M115.657866.
- Abstract
- The highly mutagenic A:oxoG (8-oxoguanine) base pair in
DNA most frequently arises by aberrant replication of the
primary oxidative lesion C:oxoG. This lesion is
particularly insidious because neither of its constituent
nucleobases faithfully transmit genetic information from
the original C:G base pair. Repair of A:oxoG is initiated
by adenine DNA glycosylase, which catalyzes hydrolytic
cleavage of the aberrant A nucleobase from the DNA
backbone. These enzymes, MutY in bacteria and MUTYH in
humans, scrupulously avoid processing of C:oxoG because
cleavage of the C residue in C:oxoG would actually promote
mutagenic conversion to A:oxoG. Here we analyze the
structural basis for rejection of C:oxoG by MutY, using a
synthetic crystallography approach to capture the enzyme in
the process of inspecting the C:oxoG anti-substrate, with
which it ordinarily binds only fleetingly. We find that
MutY uses two distinct strategies to avoid presentation of
C to the enzyme active site. Firstly, MutY possesses an
exo-site that serves as a decoy for C, and secondly,
repulsive forces with a key active site residue prevent
stable insertion of C into the nucleobase recognition
pocket within the enzyme active site.