Summary information and primary citation
- PDB-id
-
4mdf;
SNAP-derived features in text and
JSON formats
- Class
- transferase-DNA
- Method
- X-ray (1.727 Å)
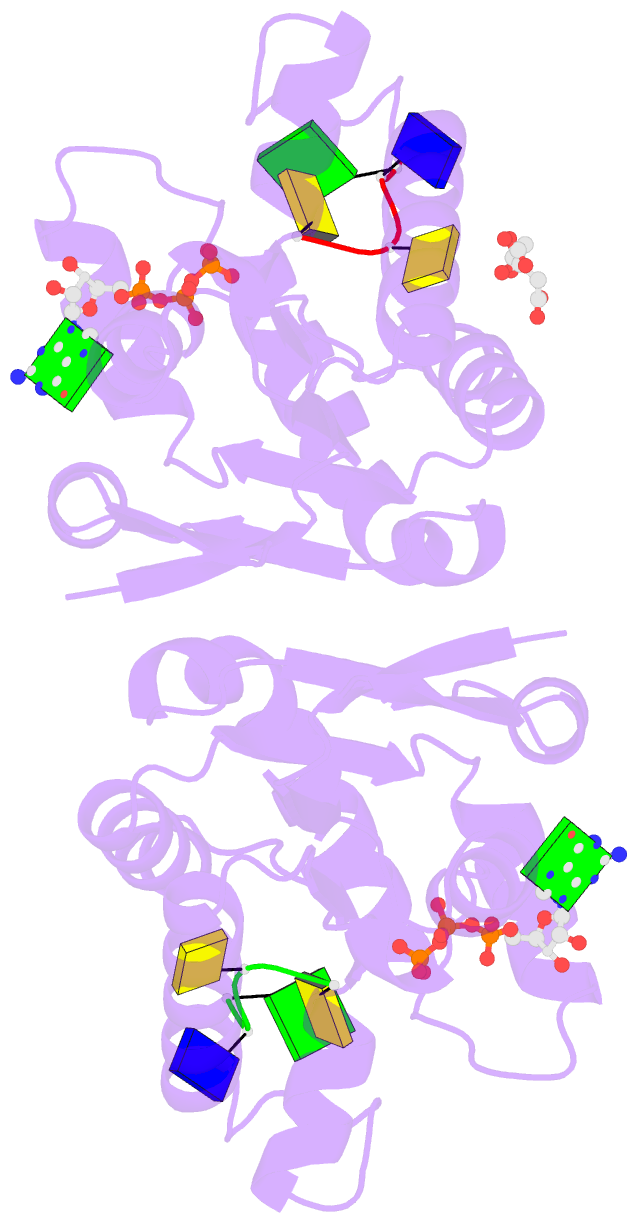
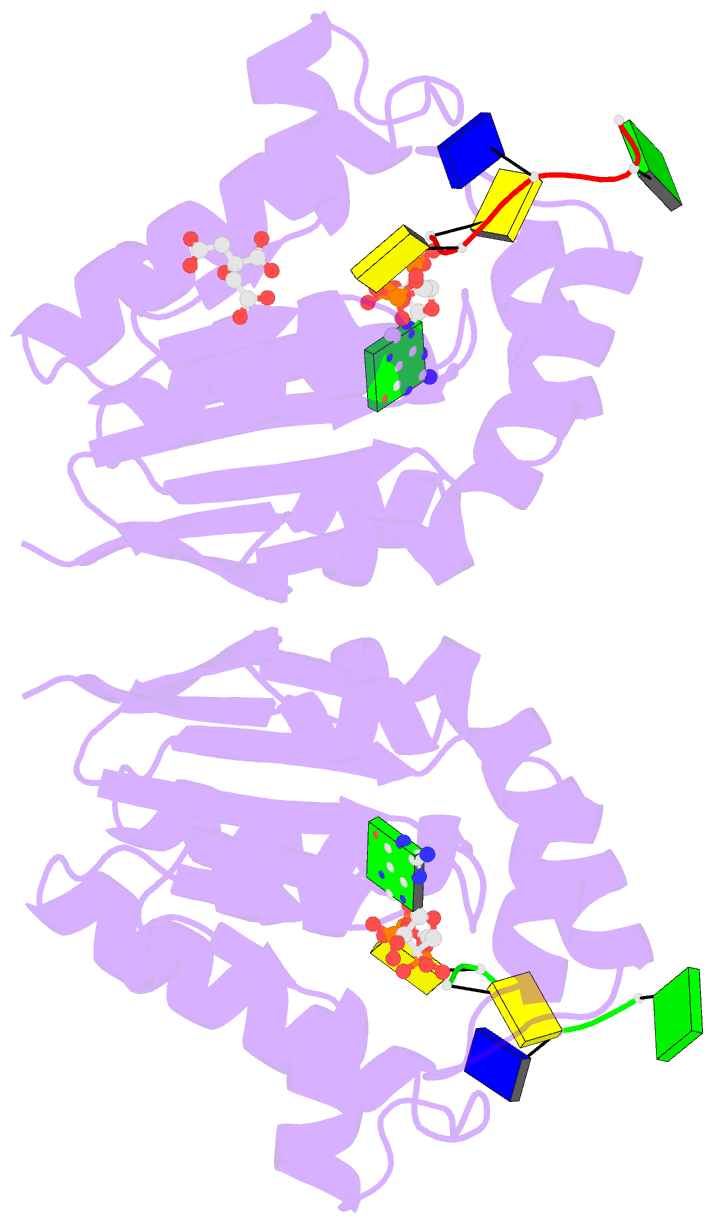
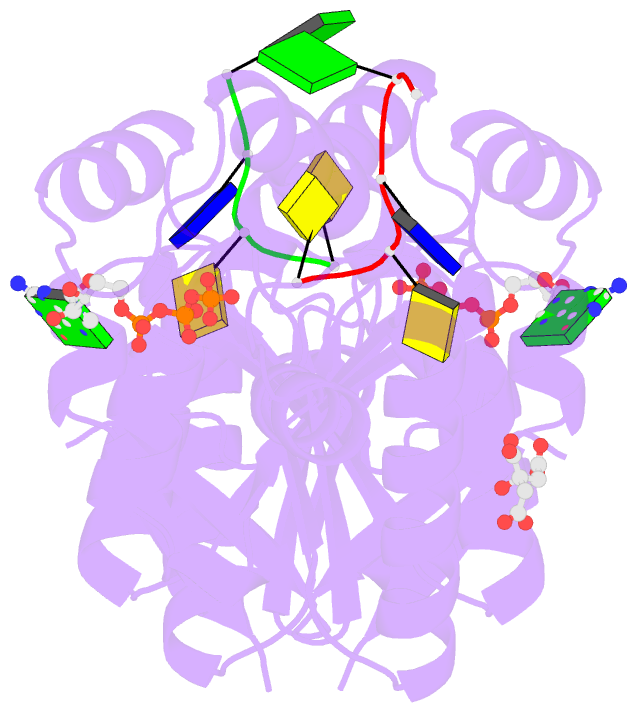
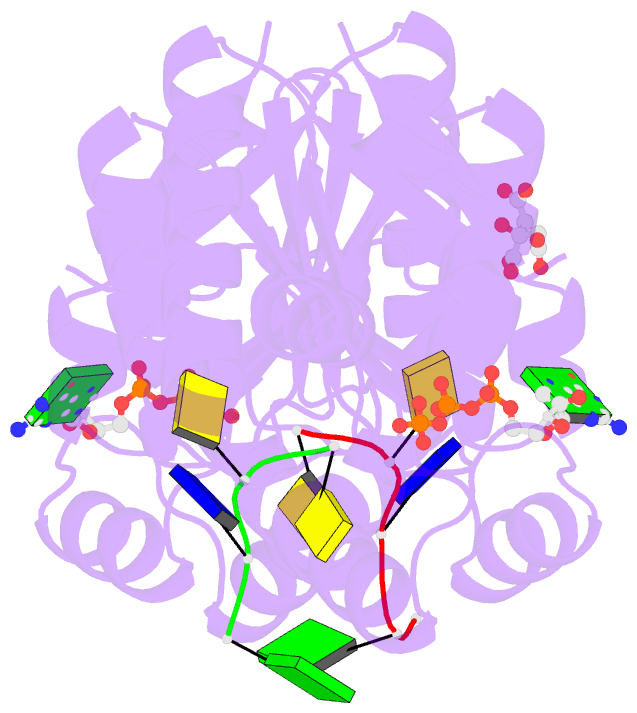
- Summary
- Structure of bacterial polynucleotide kinase michaelis
complex bound to gtp and DNA
- Reference
-
Das U, Wang LK, Smith P, Jacewicz A, Shuman S (2014):
"Structures
of bacterial polynucleotide kinase in a Michaelis complex
with GTP*Mg2+ and 5'-OH oligonucleotide and a product
complex with GDP*Mg2+ and 5'-PO4 oligonucleotide reveal a
mechanism of general acid-base catalysis and the
determinants of phosphoacceptor recognition."
Nucleic Acids Res., 42,
1152-1161. doi: 10.1093/nar/gkt936.
- Abstract
- Clostridium thermocellum polynucleotide kinase
(CthPnk), the 5' end-healing module of a bacterial RNA
repair system, catalyzes reversible phosphoryl transfer
from an NTP donor to a 5'-OH polynucleotide acceptor. Here
we report the crystal structures of CthPnk-D38N in a
Michaelis complex with GTP•Mg(2+) and a 5'-OH
oligonucleotide and a product complex with GDP•Mg(2+) and a
5'-PO4 oligonucleotide. The O5' nucleophile is situated 3.0
Å from the GTP γ phosphorus in the Michaelis complex, where
it is coordinated by Asn38 and is apical to the bridging β
phosphate oxygen of the GDP leaving group. In the product
complex, the transferred phosphate has undergone
stereochemical inversion and Asn38 coordinates the
5'-bridging phosphate oxygen of the oligonucleotide. The
D38N enzyme is poised for catalysis, but cannot execute
because it lacks Asp38-hereby implicated as the essential
general base catalyst that abstracts a proton from the
5'-OH during the kinase reaction. Asp38 serves as a general
acid catalyst during the 'reverse kinase' reaction by
donating a proton to the O5' leaving group of the 5'-PO4
strand. The acceptor strand binding mode of CthPnk is
distinct from that of bacteriophage T4 Pnk.