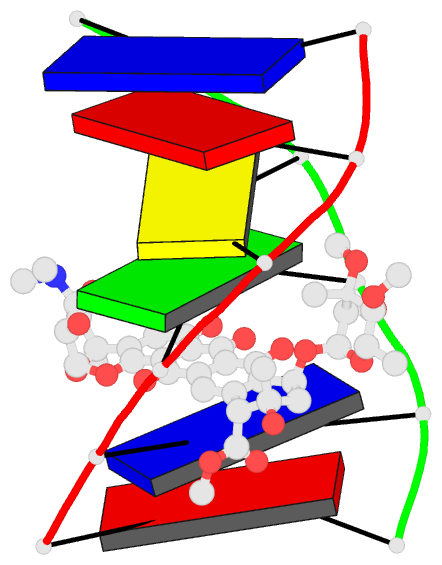
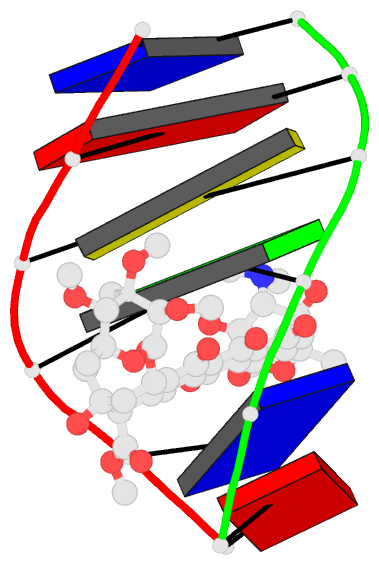
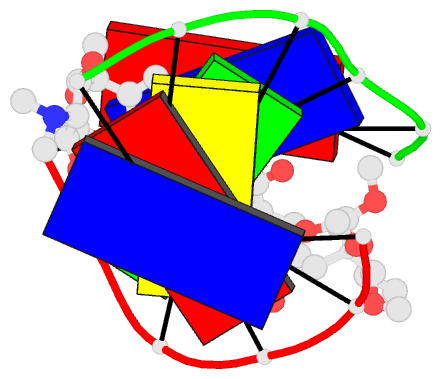
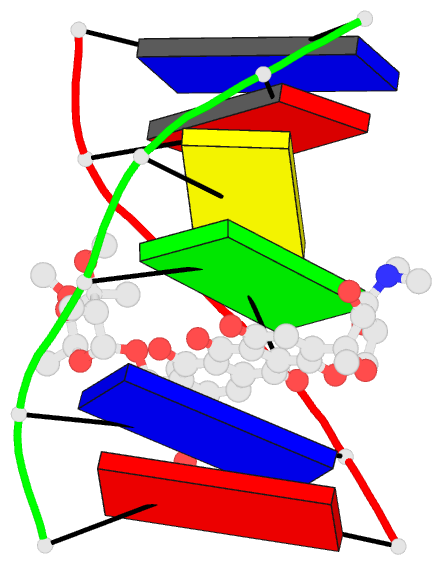
Summary information and primary citation
- PDB-id
-
1qch;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- NMR
- Summary
- Structure, dynamics and hydration of the
nogalamycin-d(atgcat)2 complex determined by NMR and
molecular dynamics simulations in solution
- Reference
-
Williams HE, Searle MS (1999): "Structure,
dynamics and hydration of the
nogalamycin-d(ATGCAT)2Complex determined by NMR and
molecular dynamics simulations in solution."
J.Mol.Biol., 290, 699-716. doi:
10.1006/jmbi.1999.2903.
- Abstract
- The structure of the 1:1 nogalamycin:d(ATGCAT)2 complex
has been determined in solution from high-resolution NMR
data and restrained molecular dynamics (rMD) simulations
using an explicit solvation model. The antibiotic
intercalates at the 5'-TpG step with the nogalose lying
along the minor groove towards the centre of the duplex.
Many drug-DNA nuclear Overhauser enhancements (NOEs) in the
minor groove are indicative of hydrophobic interactions
over the TGCA sequence. Steric occlusion prevents a second
nogalamycin molecule from binding at the symmetry-related
5'-CpA site, leading to the conclusion that the observed
binding orientation in this complex is the preferred
orientation free of the complication of end-effects (drug
molecules occupy terminal intercalation sites in all X-ray
structures) or steric interactions between drug molecules
(other NMR structures have two drug molecules bound in
close proximity), as previously suggested. Fluctuations in
key structural parameters such as rise, helical twist,
slide, shift, buckle and sugar pucker have been examined
from an analysis of the final 500 ps of a 1 ns rMD
simulation, and reveal that many sequence-dependent
structural features previously identified by comparison of
different X-ray structures lie within the range of dynamic
fluctuations observed in the MD simulations. Water density
calculations on MD simulation data reveal a time-averaged
pattern of hydration in both the major and minor groove, in
good agreement with the extensive hydration observed in two
related X-ray structures in which nogalamycin is bound at
terminal 5'-TpG sites. However, the pattern of hydration
determined from the sign and magnitude of NOE and ROE
cross-peaks to water identified in 2D NOESY and ROESY
experiments identifies only a few "bound" water molecules
with long residence times. These solvate the charged
bicycloaminoglucose sugar ring, suggesting an important
role for water molecules in mediating drug-DNA
electrostatic interactions within the major groove. The
high density of water molecules found in the minor groove
in X-ray structures and MD simulations is found to be
associated with only weakly bound solvent in solution.