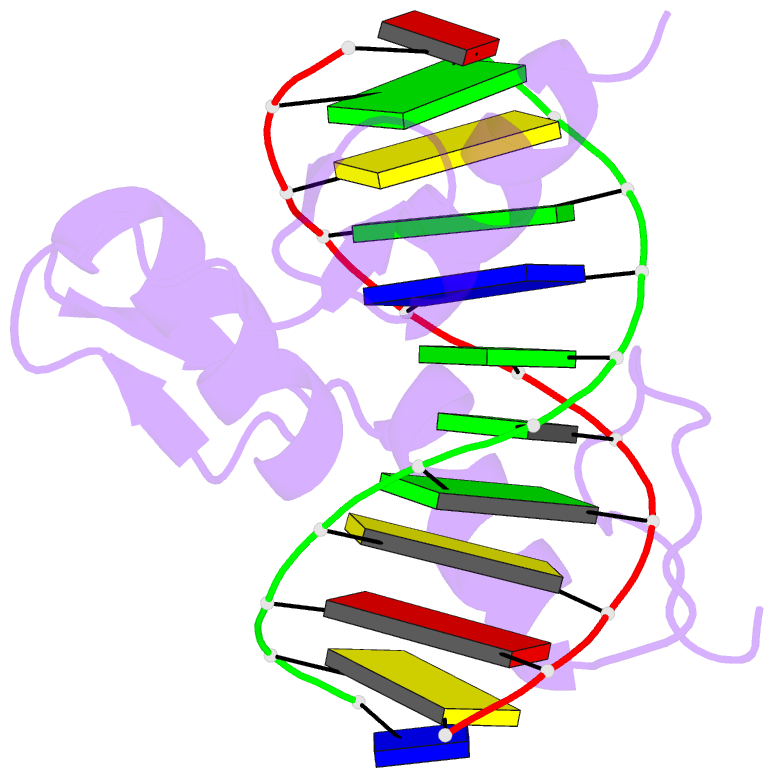
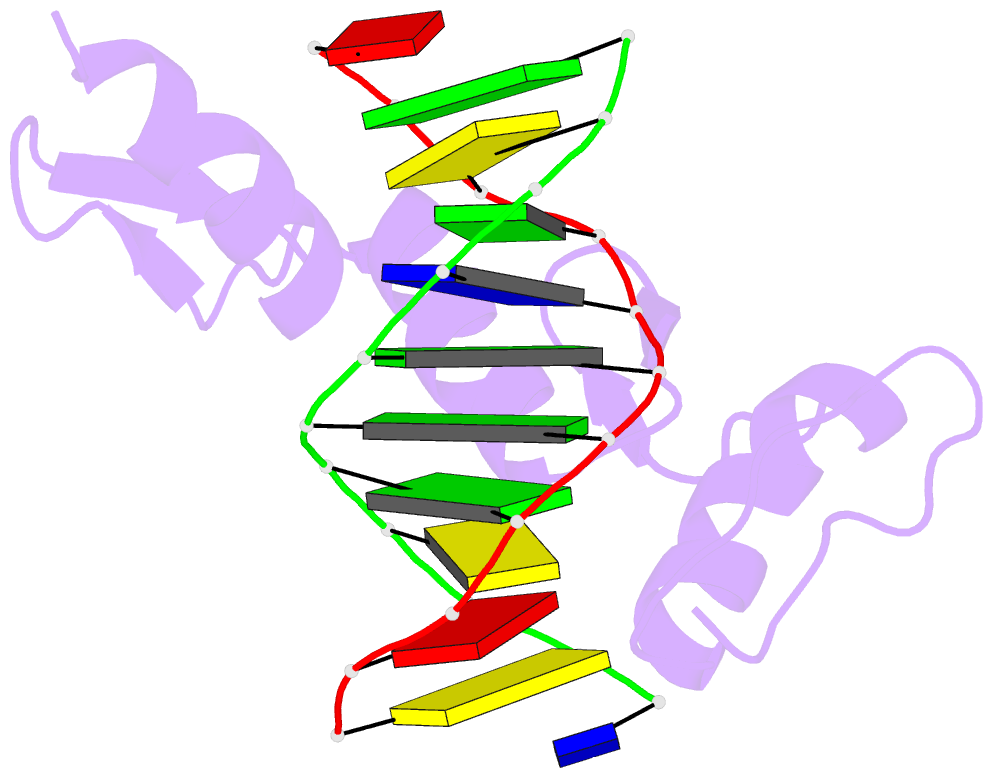
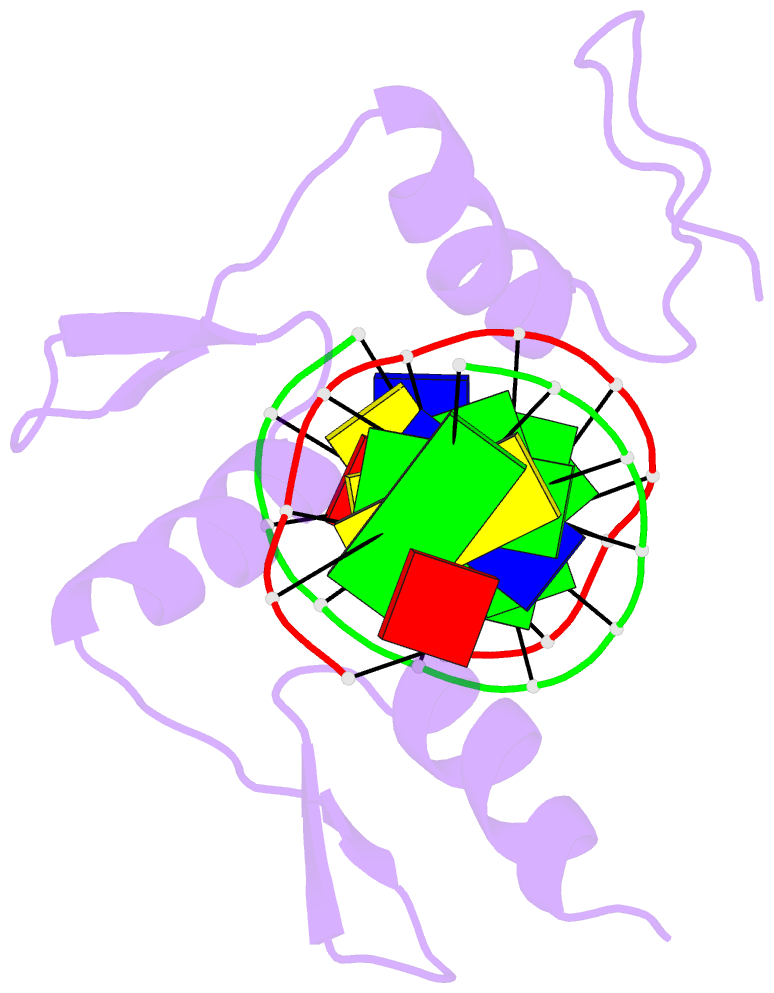
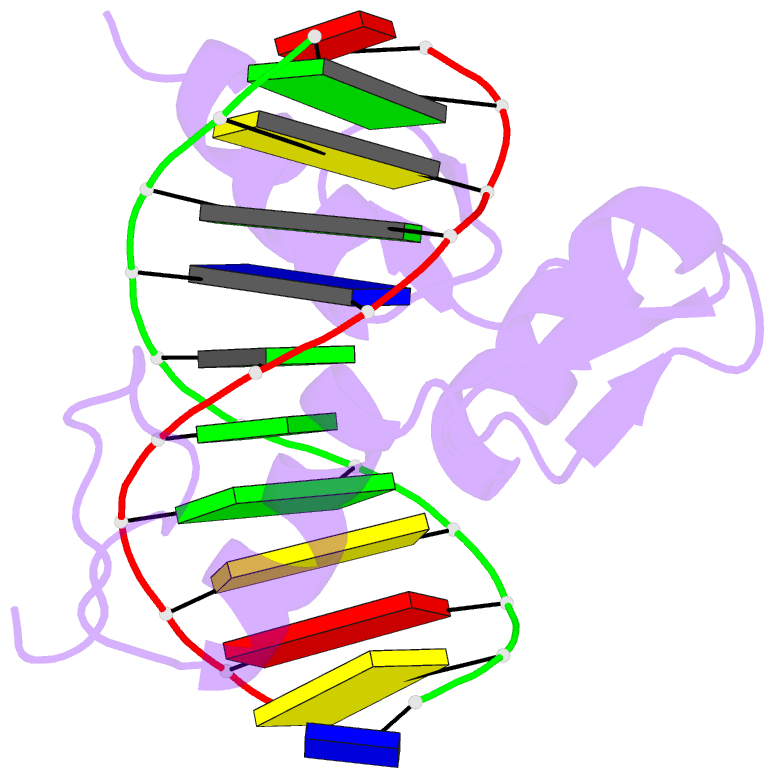
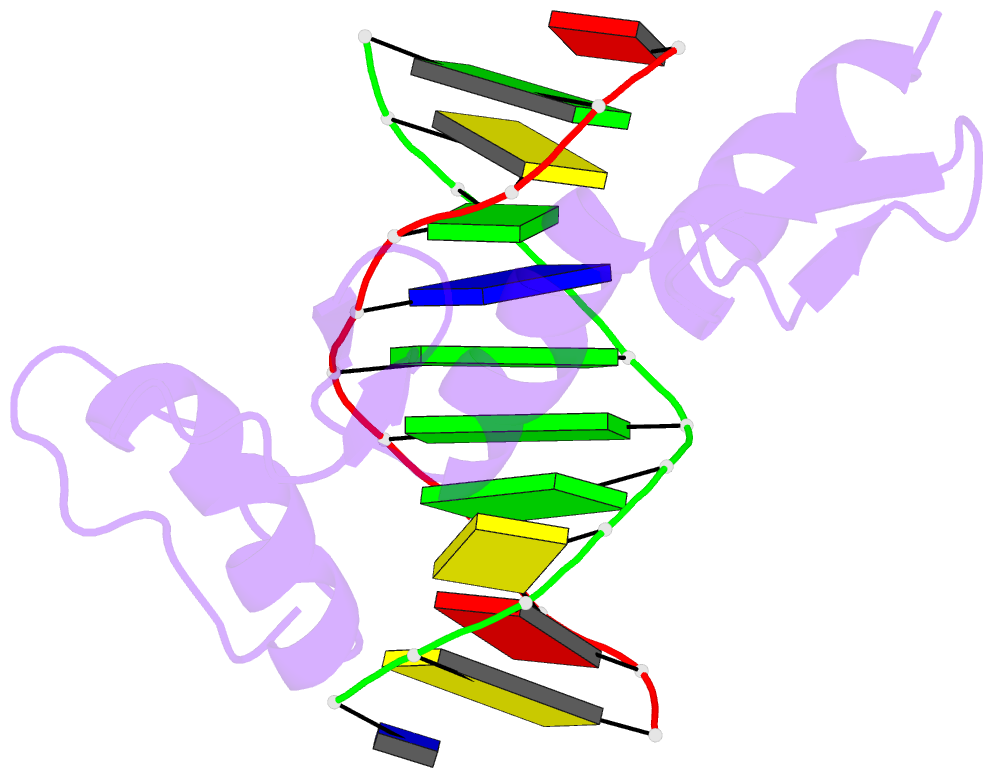
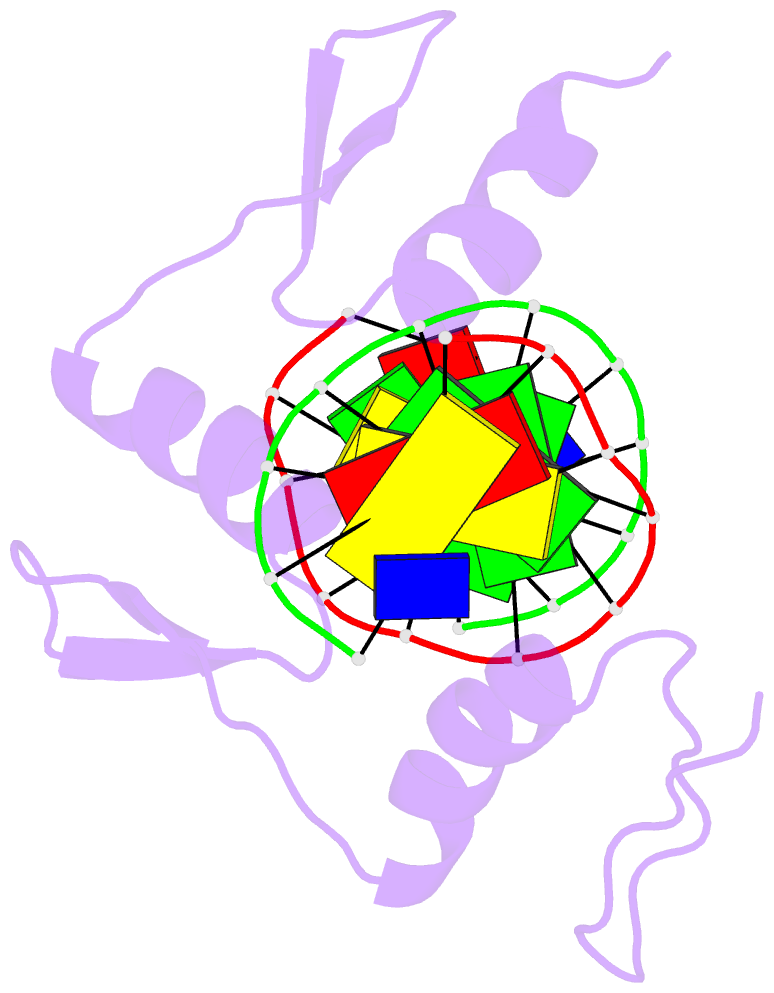
Summary information and primary citation
- PDB-id
-
1a1i;
SNAP-derived features in text and
JSON formats
- Class
- transcription-DNA
- Method
- X-ray (1.6 Å)
- Summary
- Radr (zif268 variant) zinc finger-DNA complex (gcac
site)
- Reference
-
Elrod-Erickson M, Benson TE, Pabo CO (1998): "High-resolution
structures of variant Zif268-DNA complexes: implications
for understanding zinc finger-DNA recognition."
Structure, 6, 451-464. doi:
10.1016/S0969-2126(98)00047-1.
- Abstract
- Background: Zinc fingers of the Cys2-His2 class
comprise one of the largest families of eukaryotic
DNA-binding motifs and recognize a diverse set of DNA
sequences. These proteins have a relatively simple modular
structure and key base contacts are typically made by a few
residues from each finger. These features make the zinc
finger motif an attractive system for designing novel
DNA-binding proteins and for exploring fundamental
principles of protein-DNA recognition.
Results: Here we report the X-ray crystal structures of
zinc finger-DNA complexes involving three variants of
Zif268, with multiple changes in the recognition helix of
finger one. We describe the structure of each of these
three-finger peptides bound to its corresponding target
site. To help elucidate the differential basis for
site-specific recognition, the structures of four other
complexes containing various combinations of these peptides
with alternative binding sites have also been
determined.
Conclusions: The protein-DNA contacts observed in these
complexes reveal the basis for the specificity demonstrated
by these Zif268 variants. Many, but not all, of the
contacts can be rationalized in terms of a recognition
code, but the predictive value of such a code is limited.
The structures illustrate how modest changes in the docking
arrangement accommodate the new sidechain-base and
sidechain-phosphate interactions. Such adaptations help
explain the versatility of naturally occurring zinc finger
proteins and their utility in design.