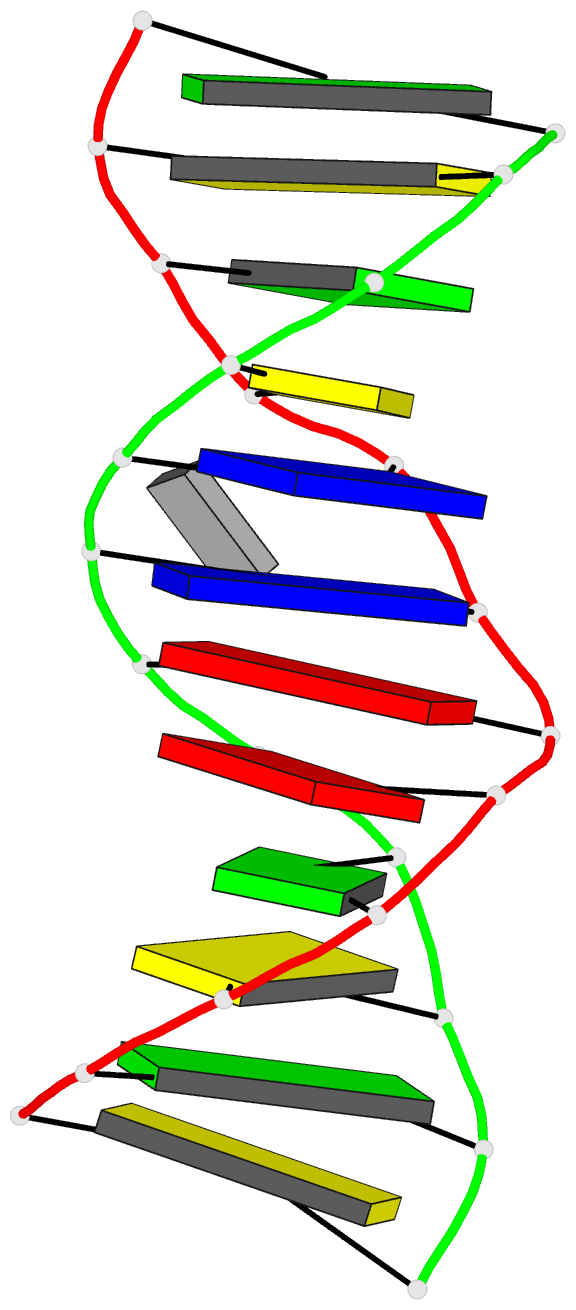
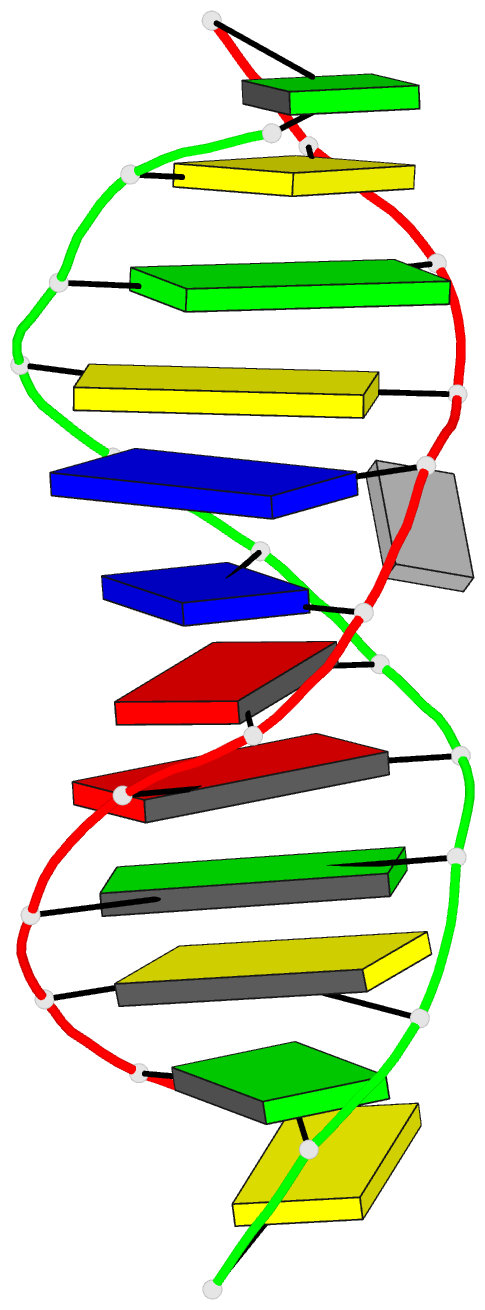
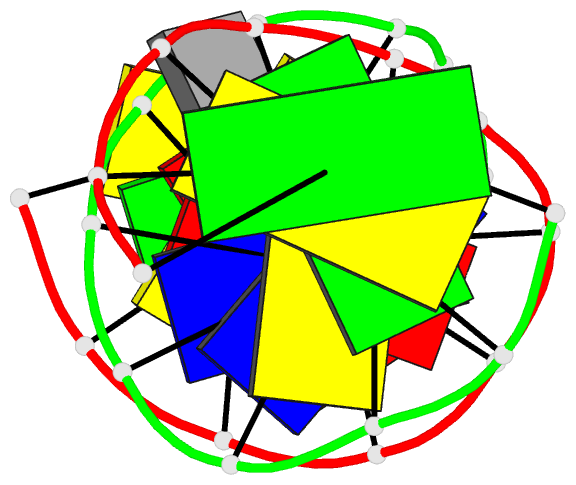
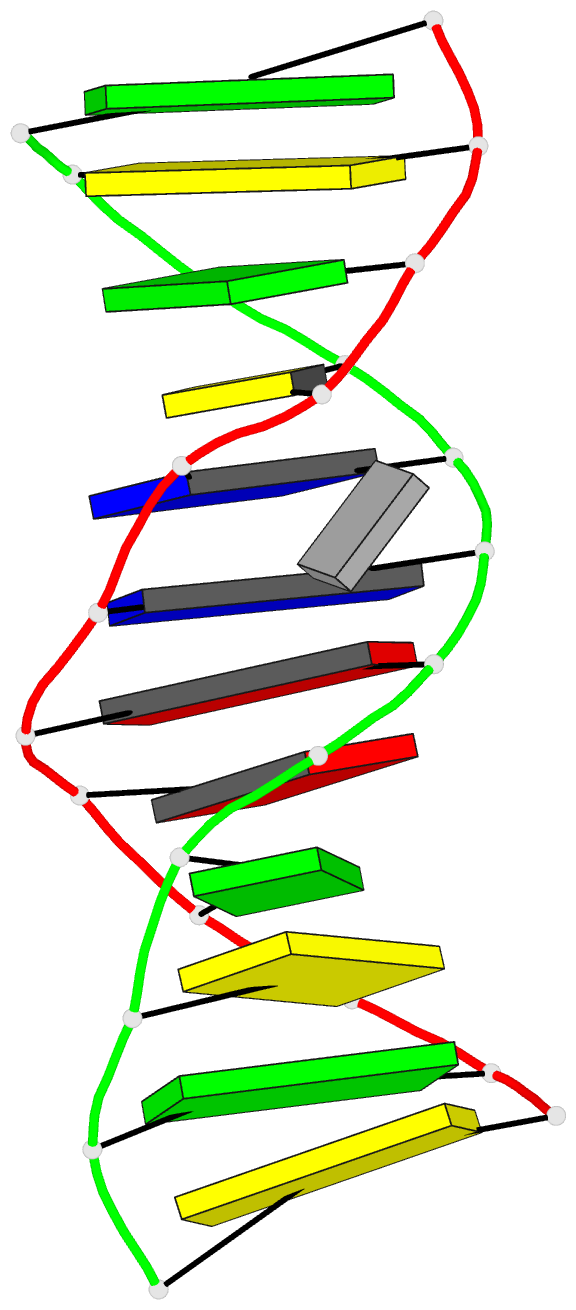
Summary information and primary citation
- PDB-id
-
144d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (2.25 Å)
- Summary
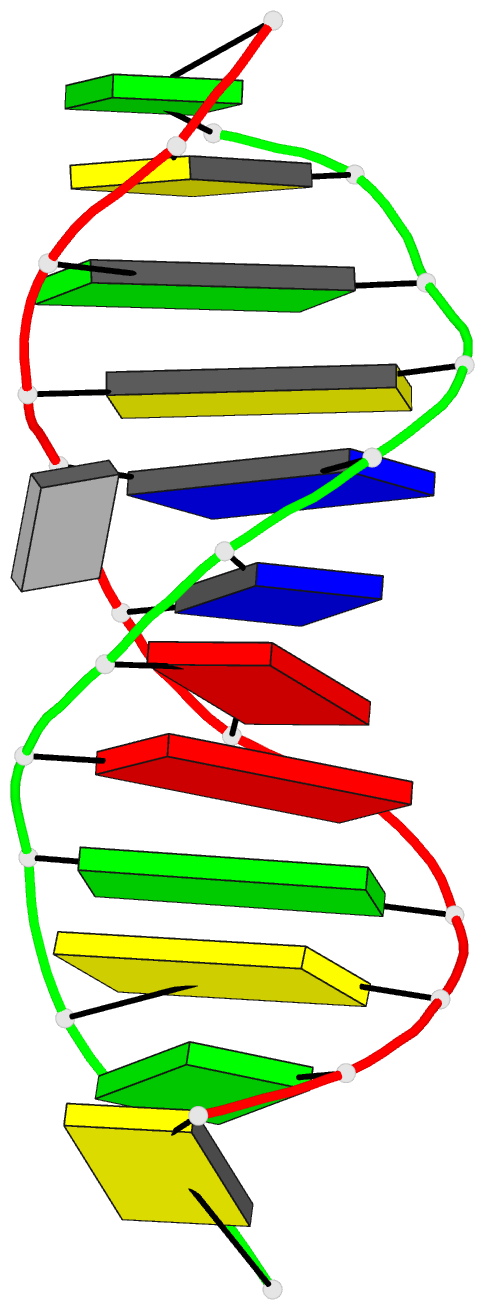
- Minor groove binding of sn6999 to an alkylated DNA:
molecular structure of d(cgc[e6g]aattcgcg)-sn6999
complex
- Reference
-
Gao YG, Sriram M, Denny WA, Wang AH (1993): "Minor
groove binding of SN6999 to an alkylated DNA: molecular
structure of d(CGC[e6G]AATTCGCG)-SN6999 complex."
Biochemistry, 32, 9639-9648.
doi: 10.1021/bi00088a016.
- Abstract
- The interaction between a potent synthetic antitumor
and antiviral minor groove binding drug
1-methyl-4-[4-[4-(4-(1-methylquinolinium)amino)benzamido]anilino]
pyridinium dichloride (SN6999) and an alkylated DNA
d(CGC[e6G]AATTCGCG) dodecamer has been studied by X-ray
crystallography. The complex forms a new crystal lattice in
the space group P2(1)2(1)2(1) with unit cell dimensions of
a = 28.48 A, b = 36.11 A, and c = 69.60 A. The structure
has been solved by the molecular replacement method and
refined to an R-factor of 17.0% at approximately 2.5 A
resolution using 1618 reflections. In the complex, the
SN6999 covers almost six base pairs in the narrow minor
groove with the 1-methylquinolinium (Q) ring near T8-A17
and the 1-methylpyridinium (P) ring near the C3-G22 base
pair. The central benzamido (BQ) and anilino (BP) rings are
essentially coplanar, with the Q and P rings having large
dihedral angles of 38 degrees and 39 degrees, respectively,
to the plane of BQ/BP. There is only one direct hydrogen
bond between the amide NH of SN6999 to T20O2 of DNA. The
drug-DNA interaction is stabilized by stacking interaction
of sugar oxygens from T20O4' to BQ and C21O4' to BP. There
is charge-induced dipole interaction between the positively
charged nitrogen atom of 1-methylquinolinium with C9O4' and
that of 1-methylpyridinium with G22O4'. The crystal
structure of the complex can be used to explain the NMR
results. SN6999 lacks the crescent shape observed in other
minor groove binding drugs and distorts the DNA duplex upon
binding. The complex packs in the lattice using the
G-N2:G-N3 interlocking base pairs at both ends of the
helix. As in earlier cases, the two independent e6G:C base
pairs adopt different base pairing schemes. The e6G16:C9
base pair adopts a previously observed bifurcated
configuration involving three-centered hydrogen bonds and
is similar to a Watson-Crick pairing. In contrast, the
e6G4:C21 base pair adopts a novel "reverse wobble"
configuration with C21 being pushed toward the major groove
side. The ethyl group is in the proximal orientation (to
N7) in both base pairs. Taken together with the
observations found in the same DNA complexed to Hoechst
33258, Hoechst 33342, and retropsin from different crystal
lattices, the results suggest that the e6G:C base pairing
is weak and polymorphic when compared to a normal G:C base
pair and the DNA duplex containing this lesion is readily
distorted.