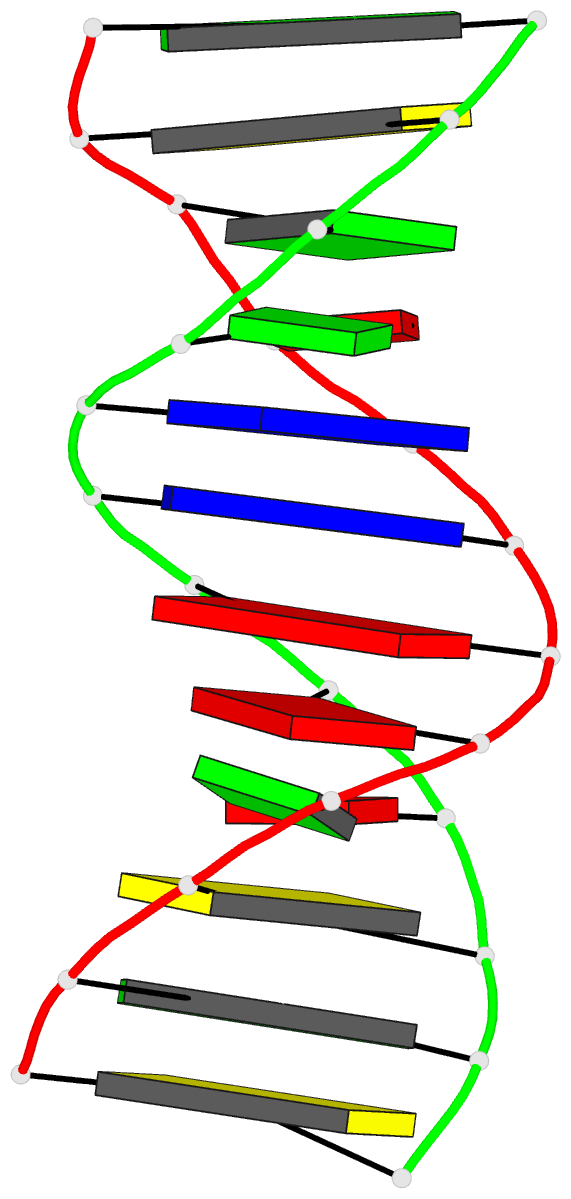
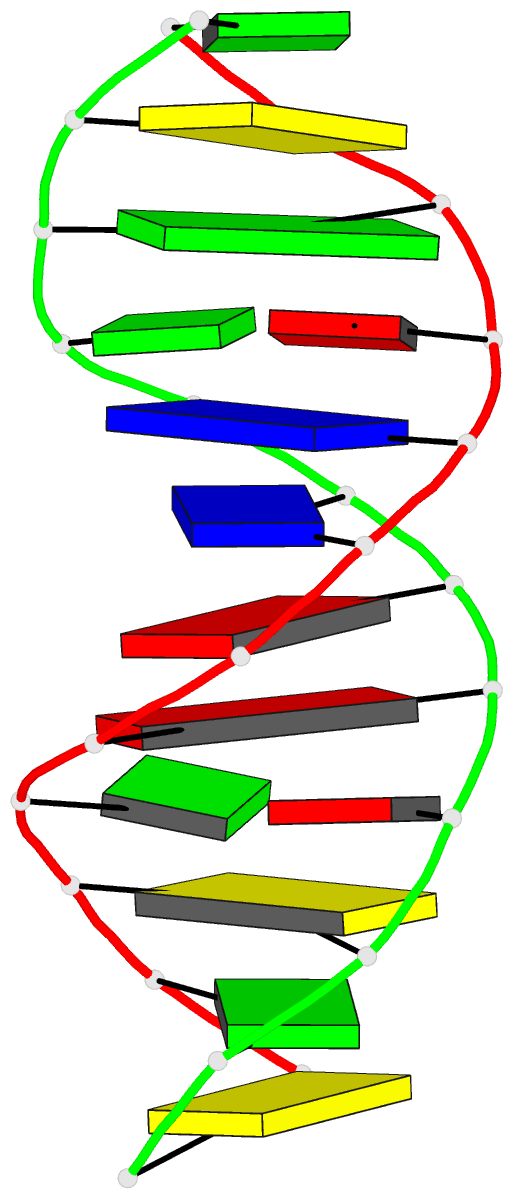
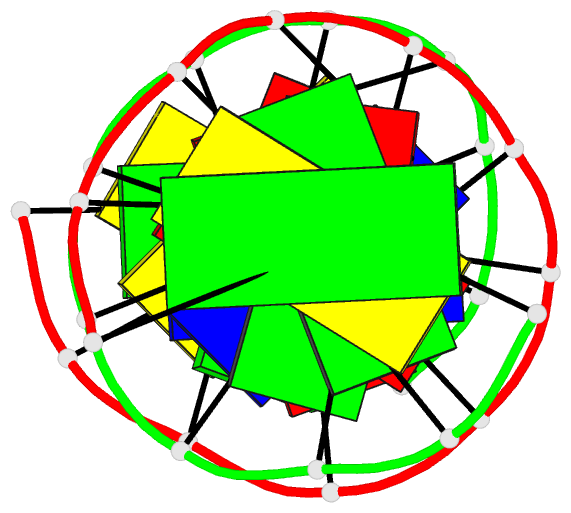
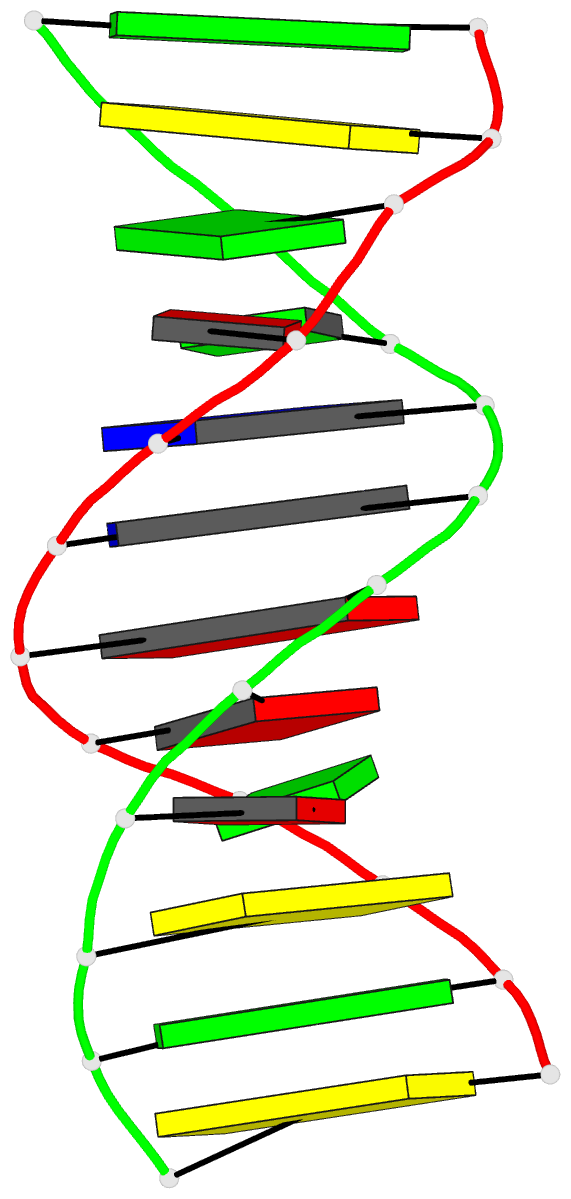
Summary information and primary citation
- PDB-id
-
112d;
SNAP-derived features in text and
JSON formats
- Class
- DNA
- Method
- X-ray (2.5 Å)
- Summary
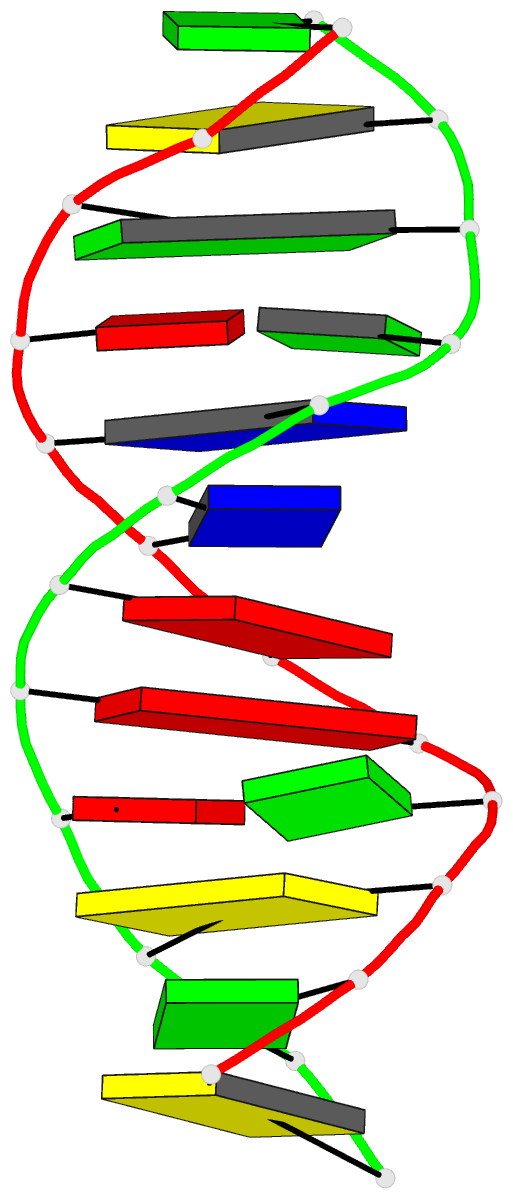
- Molecular structure of the g.a base pair in DNA and its
implications for the mechanism of transversion
mutations
- Reference
-
Brown T, Hunter WN, Kneale G, Kennard O (1986): "Molecular
structure of the G.A base pair in DNA and its
implications for the mechanism of transversion
mutations." Proc.Natl.Acad.Sci.USA,
83, 2402-2406. doi: 10.1073/pnas.83.8.2402.
- Abstract
- The synthetic deoxydodecamer d(C-G-C-G-A-A-T-T-A-G-C-G)
was analyzed by x-ray diffraction methods, and the
structure was refined to a residual error of R = 0.17 at
2.5-A resolution (2 sigma data) with 83 water molecules
located. The sequence crystallizes as a full turn of a
B-DNA helix and contains 2 purine X purine (G.A) base pairs
and 10 Watson-Crick base pairs. The analysis shows
conclusively that adenine is in the syn orientation with
respect to the sugar moiety whereas guanine adopts the
usual trans orientation. Nitrogen atoms of both bases are
involved in hydrogen bonding with the N-1 of guanine 2.84 A
from the N-7 of adenine and the N-6 of adenine within 2.74
A of the O-6 of guanine. The C-1'...C-1' separation is 10.7
A close to that for standard Watson-Crick base pairs. The
incorporation of the purine.purine base pairs at two steps
in the dodecamer causes little perturbation of either the
local or the global conformation of the double helix.
Comparison of the structural features with those of the G.T
wobble pair and the standard G.C pair suggests a rationale
for the differential enzymatic repair of the two types of
base-pair mismatches.